1JR3
 
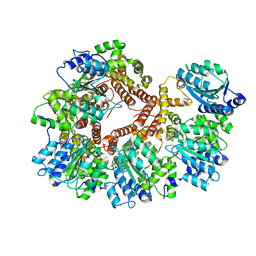 | | Crystal Structure of the Processivity Clamp Loader Gamma Complex of E. coli DNA Polymerase III | | 分子名称: | DNA polymerase III subunit gamma, DNA polymerase III, delta subunit, ... | | 著者 | Jeruzalmi, D, O'Donnell, M, Kuriyan, J. | | 登録日 | 2001-08-10 | | 公開日 | 2001-09-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of the processivity clamp loader gamma (gamma) complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JQJ
 
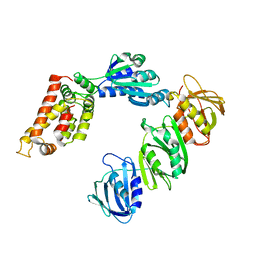 | | Mechanism of Processivity Clamp Opening by the Delta Subunit Wrench of the Clamp Loader Complex of E. coli DNA Polymerase III: Structure of the beta-delta complex | | 分子名称: | DNA polymerase III, beta chain, delta subunit | | 著者 | Jeruzalmi, D, Yurieva, O, Zhao, Y, Young, M, Stewart, J, Hingorani, M, O'Donnell, M, Kuriyan, J. | | 登録日 | 2001-08-07 | | 公開日 | 2001-11-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JQL
 
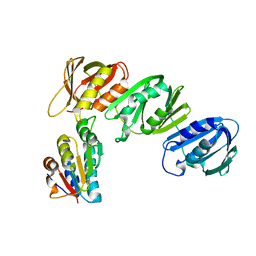 | | Mechanism of Processivity Clamp Opening by the Delta Subunit Wrench of the Clamp Loader Complex of E. coli DNA Polymerase III: Structure of beta-delta (1-140) | | 分子名称: | DNA Polymerase III, BETA CHAIN, DELTA SUBUNIT | | 著者 | Jeruzalmi, D, Yurieva, O, Zhao, Y, Young, M, Stewart, J, Hingorani, M, O'Donnell, M, Kuriyan, J. | | 登録日 | 2001-08-07 | | 公開日 | 2001-09-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
1ARO
 
 | |
7SHQ
 
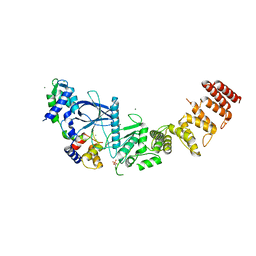 | | Structure of a functional construct of eukaryotic elongation factor 2 kinase in complex with calmodulin. | | 分子名称: | CALCIUM ION, Calmodulin-1, Eukaryotic elongation factor 2 kinase,Eukaryotic elongation factor 2 kinase, ... | | 著者 | Piserchio, A, Isiorho, E.A, Jeruzalmi, D, Dalby, K.N, Ghose, R. | | 登録日 | 2021-10-11 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structural basis for the calmodulin-mediated activation of eukaryotic elongation factor 2 kinase.
Sci Adv, 8, 2022
|
|
1QQC
 
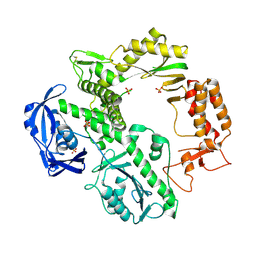 | | CRYSTAL STRUCTURE OF AN ARCHAEBACTERIAL DNA POLYMERASE D.TOK | | 分子名称: | DNA POLYMERASE II, MAGNESIUM ION, SULFATE ION | | 著者 | Zhao, Y, Jeruzalmi, D, Leighton, L, Lasken, R, Kuriyan, J. | | 登録日 | 1999-06-02 | | 公開日 | 1999-10-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of an archaebacterial DNA polymerase
Structure Fold.Des., 7, 1999
|
|
2R6F
 
 | | Crystal Structure of Bacillus stearothermophilus UvrA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Excinuclease ABC subunit A, ZINC ION | | 著者 | Inuzuka, Y, Pakotiprapha, D, Bowman, B.R, Jeruzalmi, D, Verdine, G.L. | | 登録日 | 2007-09-05 | | 公開日 | 2008-01-08 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal Structure of Bacillus stearothermophilus UvrA Provides Insight into ATP-Modulated Dimerization, UvrB Interaction, and DNA Binding.
Mol.Cell, 29, 2008
|
|
6BBM
 
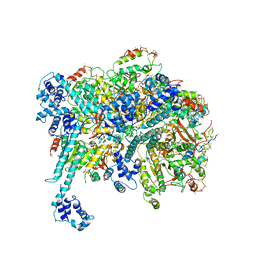 | | Mechanisms of Opening and Closing of the Bacterial Replicative Helicase: The DnaB Helicase and Lambda P Helicase Loader Complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Replication protein P, Replicative DNA helicase | | 著者 | Chase, J, Catalano, A, Noble, A.J, Eng, E.T, Olinares, P.D.B, Molloy, K, Pakotiprapha, D, Samuels, M, Chain, B, des Georges, A, Jeruzalmi, D. | | 登録日 | 2017-10-18 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Mechanisms of opening and closing of the bacterial replicative helicase.
Elife, 7, 2018
|
|
3FPN
 
 | |
6N9L
 
 | | Crystal structure of T. maritima UvrA d117-399 with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, UvrABC system protein A, ZINC ION | | 著者 | Hartley, S, Case, B, Osuga, M, Hingorani, M.M, Jeruzalmi, D. | | 登録日 | 2018-12-03 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | The ATPase mechanism of UvrA2 reveals the distinct roles of proximal and distal ATPase sites in nucleotide excision repair.
Nucleic Acids Res., 47, 2019
|
|
3UWX
 
 | |
3UX8
 
 | | Crystal structure of UvrA | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Excinuclease ABC, A subunit, ... | | 著者 | Samuels, M.A, Pakotiprapha, D, Jeruzalmi, D. | | 登録日 | 2011-12-04 | | 公開日 | 2012-02-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and mechanism of the UvrA-UvrB DNA damage sensor.
Nat.Struct.Mol.Biol., 19, 2012
|
|
1D5A
 
 | | CRYSTAL STRUCTURE OF AN ARCHAEBACTERIAL DNA POLYMERASE D.TOK. DEPOSITION OF SECOND NATIVE STRUCTURE AT 2.4 ANGSTROM | | 分子名称: | MAGNESIUM ION, PROTEIN (DNA POLYMERASE), SULFATE ION | | 著者 | Zhao, Y, Jeruzalmi, D, Leighton, L, Lasken, R, Kuriyan, J. | | 登録日 | 1999-10-06 | | 公開日 | 2000-03-24 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of an archaebacterial DNA polymerase.
Structure Fold.Des., 7, 1999
|
|
1CEZ
 
 | |
5UBE
 
 | |
5UBF
 
 | |
5UBD
 
 | | Crystal structure of the N-terminal domain (domain 1) of RctB, RctB-1-124-L48M | | 分子名称: | RctB replication initiator protein | | 著者 | Orlova, N, Ivashkiv, O, Waldor, M.K, Jeruzalmi, D. | | 登録日 | 2016-12-20 | | 公開日 | 2017-01-11 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | The replication initiator of the cholera pathogen's second chromosome shows structural similarity to plasmid initiators.
Nucleic Acids Res., 45, 2017
|
|
1BF5
 
 | | TYROSINE PHOSPHORYLATED STAT-1/DNA COMPLEX | | 分子名称: | DNA (5'-D(*AP*CP*AP*GP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*G P*C)-3'), DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*AP*CP*GP*GP*GP*AP*AP*AP*CP*T P*G)-3'), SIGNAL TRANSDUCER AND ACTIVATOR OF TRANSCRIPTION 1-ALPHA/BETA | | 著者 | Kuriyan, J, Zhao, Y, Chen, X, Vinkemeier, U, Jeruzalmi, D, Darnell Jr, J.E. | | 登録日 | 1998-05-27 | | 公開日 | 1998-08-12 | | 最終更新日 | 2022-12-21 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA.
Cell(Cambridge,Mass.), 93, 1998
|
|
1CZD
 
 | | CRYSTAL STRUCTURE OF THE PROCESSIVITY CLAMP GP45 FROM BACTERIOPHAGE T4 | | 分子名称: | DNA POLYMERASE ACCESSORY PROTEIN G45 | | 著者 | Moarefi, I, Jeruzalmi, D, Turner, J, O'Donnell, M, Kuriyan, J. | | 登録日 | 1999-09-02 | | 公開日 | 2000-03-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage.
J.Mol.Biol., 296, 2000
|
|
3HFH
 
 | | Crystal structure of tandem FF domains | | 分子名称: | Transcription elongation regulator 1 | | 著者 | Lu, M, Yang, J, Ren, Z, Subir, S, Bedford, M.T, Jacobson, R.H, McMurray, J.S, Chen, X. | | 登録日 | 2009-05-11 | | 公開日 | 2009-08-18 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.703 Å) | | 主引用文献 | Crystal Structure of the Three Tandem FF Domains of the Transcription Elongation Regulator CA150.
J.Mol.Biol., 393, 2009
|
|
