5AWF
 
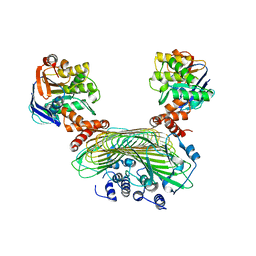 | | Crystal structure of SufB-SufC-SufD complex from Escherichia coli | | 分子名称: | FeS cluster assembly protein SufB, FeS cluster assembly protein SufD, Probable ATP-dependent transporter SufC | | 著者 | Hirabayashi, K, Wada, K. | | 登録日 | 2015-07-03 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.957 Å) | | 主引用文献 | Functional Dynamics Revealed by the Structure of the SufBCD Complex, a Novel ATP-binding Cassette (ABC) Protein That Serves as a Scaffold for Iron-Sulfur Cluster Biogenesis
J.Biol.Chem., 290, 2015
|
|
5AWG
 
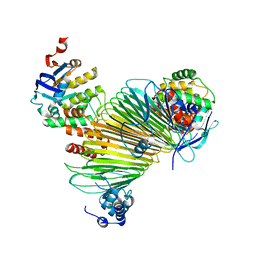 | |
5ZHS
 
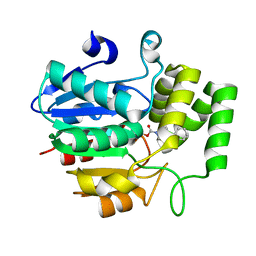 | | Crystal structure of OsD14 in complex with covalently bound KK052 | | 分子名称: | (4-phenylpiperazin-1-yl)(1H-1,2,3-triazol-1-yl)methanone, Strigolactone esterase D14 | | 著者 | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | 登録日 | 2018-03-13 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
5ZHT
 
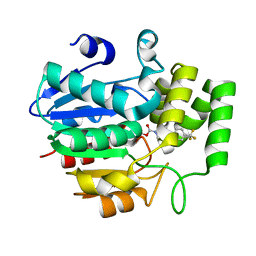 | | Crystal structure of OsD14 in complex with covalently bound KK073 | | 分子名称: | (1H-1,2,3-triazol-1-yl){4-[4-(trifluoromethyl)phenyl]piperazin-1-yl}methanone, Strigolactone esterase D14 | | 著者 | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | 登録日 | 2018-03-13 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.532 Å) | | 主引用文献 | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
5ZHR
 
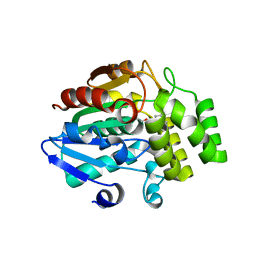 | | Crystal structure of OsD14 in complex with covalently bound KK094 | | 分子名称: | (2,3-dihydro-1H-indol-1-yl)(1H-1,2,3-triazol-1-yl)methanone, Strigolactone esterase D14 | | 著者 | Hirabayashi, K, Miyakawa, T, Tanokura, M. | | 登録日 | 2018-03-13 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Triazole Ureas Covalently Bind to Strigolactone Receptor and Antagonize Strigolactone Responses.
Mol Plant, 12, 2019
|
|
5YZ7
 
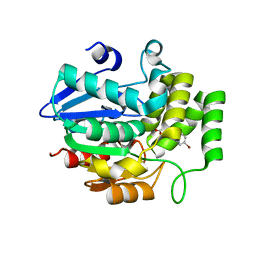 | | Crystal structure of OsD14 in complex with D-ring-opened 7'-carba-4BD | | 分子名称: | (2Z,4S)-5-(4-bromophenyl)-4-hydroxy-2-methylpent-2-enoic acid, Strigolactone esterase D14 | | 著者 | Hirabayashi, K, Jiang, K, Xu, Y, Miyakawa, T, Asami, T, Tanokura, M. | | 登録日 | 2017-12-13 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | Rationally Designed Strigolactone Analogs as Antagonists of the D14 Receptor.
Plant Cell Physiol., 59, 2018
|
|
7CD0
 
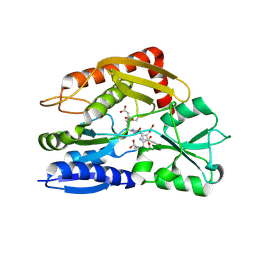 | | Crystal structure of the 2-iodoporphobilinogen-bound ES2 intermediate form of human hydroxymethylbilane synthase | | 分子名称: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, 3-[5-(aminomethyl)-4-(carboxymethyl)-2-iodo-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | 著者 | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | 登録日 | 2020-06-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
7CCZ
 
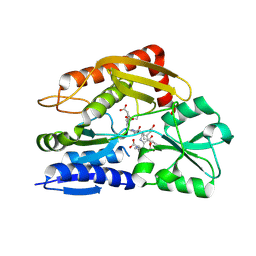 | | Crystal structure of the ES2 intermediate form of human hydroxymethylbilane synthase | | 分子名称: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | 著者 | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | 登録日 | 2020-06-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
7CCX
 
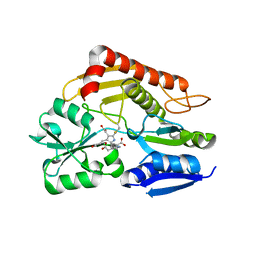 | | Crystal structure of the holo form of human hydroxymethylbilane synthase | | 分子名称: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | 著者 | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | 登録日 | 2020-06-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
7CCY
 
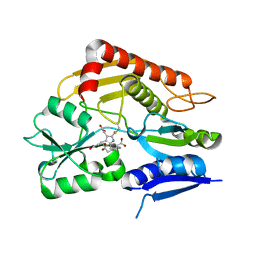 | | Crystal structure of the 2-iodoporphobilinogen-bound holo form of human hydroxymethylbilane synthase | | 分子名称: | 3-[5-(aminomethyl)-4-(carboxymethyl)-2-iodo-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | 著者 | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | 登録日 | 2020-06-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
3AV8
 
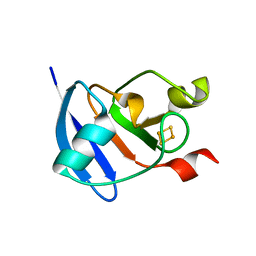 | | Refined Structure of Plant-type [2Fe-2S] Ferredoxin I from Aphanothece sacrum at 1.46 A Resolution | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin-1, SULFATE ION | | 著者 | Kameda, H, Hirabayashi, K, Wada, K, Fukuyama, K. | | 登録日 | 2011-02-28 | | 公開日 | 2012-01-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Mapping of protein-protein interaction sites in the plant-type [2Fe-2S] ferredoxin.
Plos One, 6, 2011
|
|
5ZD4
 
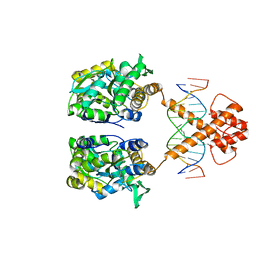 | | Crystal structure of MBP-fused BIL1/BZR1 in complex with double-stranded DNA | | 分子名称: | 1,2-ETHANEDIOL, DNA (5'-D(*TP*TP*CP*AP*CP*AP*CP*GP*TP*GP*TP*GP*AP*AP*A)-3'), Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1, ... | | 著者 | Nosaki, S, Miyakawa, T, Xu, Y, Nakamura, A, Hirabayashi, K, Tanokura, M. | | 登録日 | 2018-02-22 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structural basis for brassinosteroid response by BIL1/BZR1.
Nat Plants, 4, 2018
|
|
4PAE
 
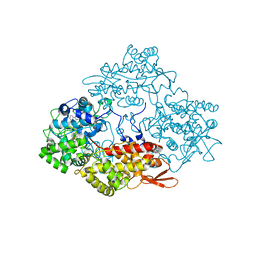 | | Crystal structure of catalase-peroxidase (KatG) W78F mutant from Synechococcus elongatus PCC7942 | | 分子名称: | Catalase-peroxidase, HEME B/C, SODIUM ION, ... | | 著者 | Kamachi, S, Wada, K, Tada, T. | | 登録日 | 2014-04-08 | | 公開日 | 2015-01-14 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.206 Å) | | 主引用文献 | Crystal structure of the catalase-peroxidase KatG W78F mutant from Synechococcus elongatus PCC7942 in complex with the antitubercular pro-drug isoniazid.
Febs Lett., 589, 2015
|
|
8K5H
 
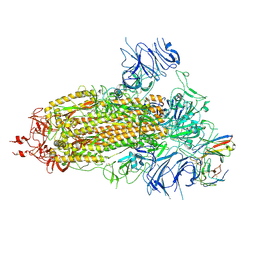 | | Structure of the SARS-CoV-2 BA.1 spike with UT28-RD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Chen, L, Kita, S, Anraku, Y, Maenaka, K. | | 登録日 | 2023-07-21 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Rational in silico design identifies two mutations that restore UT28K SARS-CoV-2 monoclonal antibody activity against Omicron BA.1.
Structure, 32, 2024
|
|
8K5G
 
 | | Structure of the SARS-CoV-2 BA.1 RBD with UT28-RD | | 分子名称: | Spike protein S1, UT28K-RD Fab Heavy chain, UT28K-RD Fab Light chain | | 著者 | Chen, L, Kita, S, Anraku, Y, Maenaka, K. | | 登録日 | 2023-07-21 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Rational in silico design identifies two mutations that restore UT28K SARS-CoV-2 monoclonal antibody activity against Omicron BA.1.
Structure, 32, 2024
|
|
5B3T
 
 | |
5B3V
 
 | |
5B3U
 
 | |
3WXO
 
 | | Crystal structure of isoniazid bound KatG catalase peroxidase from Synechococcus elongatus PCC7942 | | 分子名称: | Catalase-peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | 著者 | Wada, K, Tada, T, Kamachi, S. | | 登録日 | 2014-08-04 | | 公開日 | 2015-01-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | The crystal structure of isoniazid-bound KatG catalase-peroxidase from Synechococcus elongatus PCC7942.
Febs J., 282, 2015
|
|
3X16
 
 | |
