9CIJ
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/L36R at cryogenic temperature | | 分子名称: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | 著者 | Zhang, Y, Schlessman, L.J, Siegler, M.A, Garcia-Moreno E, B. | | 登録日 | 2024-07-03 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23E/L36R at cryogenic temperature
To Be Published
|
|
9CIK
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23R/L36D at cryogenic temperature | | 分子名称: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | 著者 | Zhang, Y, Schlessman, J.L, SIegler, M.A, Garcia-Moreno E, B. | | 登録日 | 2024-07-03 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23R/L36D at cryogenic temperature
To Be Published
|
|
9CU9
 
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D/L36K at cryogenic temperature | | 分子名称: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | 著者 | Zhang, Y, Schlessman, J.L, Robinson, A.C, Garcia-Moreno E, B. | | 登録日 | 2024-07-26 | | 公開日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of Staphylococcal nuclease variant Delta+PHS V23D/L36K at cryogenic temperature
To Be Published
|
|
5M5G
 
 | | Crystal structure of the Chaetomium Thermophilum polycomb repressive complex 2 (PRC2) | | 分子名称: | Fragment from molecular 2 (region containing putative polycomb protein Suz12), HISTONE H3 11-Mer peptide, Putative uncharacterized protein, ... | | 著者 | Zhang, Y, Justin, N, Wilson, J, Gamblin, S. | | 登録日 | 2016-10-21 | | 公開日 | 2017-01-11 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Comment on "Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2".
Science, 354, 2016
|
|
3RC0
 
 | | Human SETD6 in complex with RelA Lys310 peptide | | 分子名称: | 1,2-ETHANEDIOL, N-lysine methyltransferase SETD6, S-ADENOSYLMETHIONINE, ... | | 著者 | Chang, Y, Levy, D, Horton, J.R, Peng, J, Zhang, X, Gozani, O, Cheng, X. | | 登録日 | 2011-03-30 | | 公開日 | 2011-05-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling.
Nucleic Acids Res., 39, 2011
|
|
2L3M
 
 | | Solution structure of the putative copper-ion-binding protein from Bacillus anthracis str. Ames | | 分子名称: | Copper-ion-binding protein | | 著者 | Zhang, Y, Winsor, J, Dubrovska, I, Anderson, W, Radhakrishnan, I, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2010-09-16 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | To be published
To be Published
|
|
2LD5
 
 | |
2A9Z
 
 | | Crystal structure of T. gondii adenosine kinase complexed with N6-dimethyladenosine and AMP-PCP | | 分子名称: | 6N-DIMETHYLADENOSINE, CHLORIDE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | 著者 | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | 登録日 | 2005-07-12 | | 公開日 | 2006-07-11 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2A9Y
 
 | | Crystal structure of T. gondii adenosine kinase complexed with N6-dimethyladenosine | | 分子名称: | 6N-DIMETHYLADENOSINE, ACETATE ION, CHLORIDE ION, ... | | 著者 | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | 登録日 | 2005-07-12 | | 公開日 | 2006-07-11 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2AA0
 
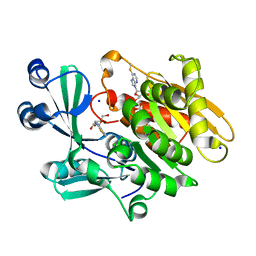 | | Crystal structure of T. gondii adenosine kinase complexed with 6-methylmercaptopurine riboside | | 分子名称: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | 登録日 | 2005-07-13 | | 公開日 | 2006-07-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2AB8
 
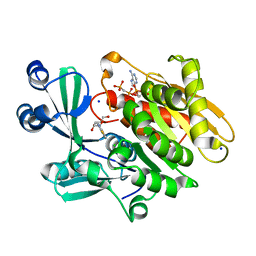 | | Crystal structure of T. gondii adenosine kinase complexed with 6-methylmercaptopurine riboside and AMP-PCP | | 分子名称: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, CHLORIDE ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | 著者 | Zhang, Y, el Kouni, M.H, Ealick, S.E. | | 登録日 | 2005-07-14 | | 公開日 | 2006-07-25 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Substrate analogs induce an intermediate conformational change in Toxoplasma gondii adenosine kinase
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2A7Q
 
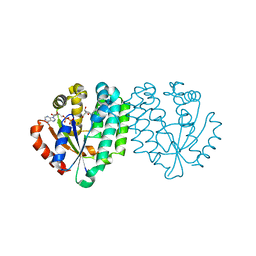 | | Crystal structure of human dCK complexed with clofarabine and ADP | | 分子名称: | 2-CHLORO-9-(2-DEOXY-2-FLUORO-B -D-ARABINOFURANOSYL)-9H-PURIN-6-AMINE, ADENOSINE-5'-DIPHOSPHATE, Deoxycytidine kinase, ... | | 著者 | Zhang, Y, Secrist III, J.A, Ealick, S.E. | | 登録日 | 2005-07-05 | | 公開日 | 2006-01-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | The structure of human deoxycytidine kinase in complex with clofarabine reveals key interactions for prodrug activation.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
8XRY
 
 | |
8XNG
 
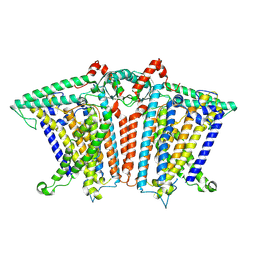 | |
8XS4
 
 | |
8XW1
 
 | |
8XW2
 
 | |
8XW3
 
 | |
8XS5
 
 | |
8XW0
 
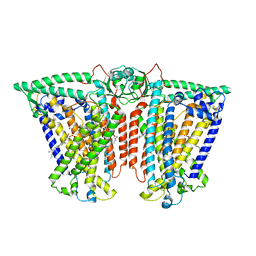 | | Cryo-EM structure of OSCA3.1-GDN state | | 分子名称: | CSC1-like protein ERD4, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | 著者 | Zhang, Y, Han, Y. | | 登録日 | 2024-01-15 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.11 Å) | | 主引用文献 | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XVY
 
 | |
8XVZ
 
 | |
8XW4
 
 | |
8XS0
 
 | |
8XVX
 
 | |
