2A5M
 
 | |
2K4C
 
 | | tRNAPhe-based homology model for tRNAVal refined against base N-H RDCs in two media and SAXS data | | 分子名称: | 76-MER | | 著者 | Grishaev, A, Ying, J, Canny, M.D, Pardi, A, Bax, A. | | 登録日 | 2008-06-04 | | 公開日 | 2008-12-09 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Solution structure of tRNAVal from refinement of homology model against residual dipolar coupling and SAXS data.
J.Biomol.Nmr, 42, 2008
|
|
2JQX
 
 | | Solution structure of Malate Synthase G from joint refinement against NMR and SAXS data | | 分子名称: | Malate synthase G | | 著者 | Grishaev, A, Tugarinov, V, Kay, L.E, Trewhella, J, Bax, A. | | 登録日 | 2007-06-13 | | 公開日 | 2007-07-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Refined solution structure of the 82-kDa enzyme malate synthase G from joint NMR and synchrotron SAXS restraints
J.Biomol.Nmr, 40, 2008
|
|
2N7J
 
 | |
4D1Q
 
 | | Hermes transposase bound to its terminal inverted repeat | | 分子名称: | SODIUM ION, TERMINAL INVERTED REPEAT, TRANSPOSASE | | 著者 | Hickman, A.B, Ewis, H, Li, X, Knapp, J, Laver, T, Doss, A.L, Tolun, G, Steven, A, Grishaev, A, Bax, A, Atkinson, P, Craig, N.L, Dyda, F. | | 登録日 | 2014-05-04 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.4 Å) | | 主引用文献 | Structural Basis of Hat Transposon End Recognition by Hermes, an Octameric DNA Transposase from Musca Domestica.
Cell(Cambridge,Mass.), 158, 2014
|
|
5I1R
 
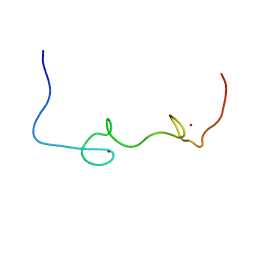 | | Quantitative characterization of configurational space sampled by HIV-1 nucleocapsid using solution NMR and X-ray scattering | | 分子名称: | Nucleocapsid protein p7, ZINC ION | | 著者 | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G.M. | | 登録日 | 2016-02-05 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Quantitative Characterization of Configurational Space Sampled by HIV-1 Nucleocapsid Using Solution NMR, X-ray Scattering and Protein Engineering.
Chemphyschem, 17, 2016
|
|
2XDF
 
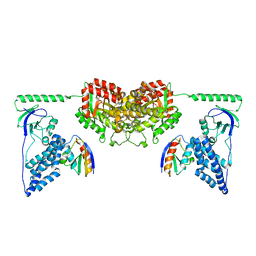 | | Solution Structure of the Enzyme I Dimer Complexed with HPr Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | 分子名称: | PHOSPHOCARRIER PROTEIN HPR, PHOSPHOENOLPYRUVATE-PROTEIN PHOSPHOTRANSFERASE | | 著者 | Schwieters, C.D, Suh, J.-Y, Grishaev, A, Guirlando, R, Takayama, Y, Clore, G.M. | | 登録日 | 2010-04-30 | | 公開日 | 2010-09-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Solution Structure of the 128 kDa Enzyme I Dimer from Escherichia Coli and its 146 kDa Complex with Hpr Using Residual Dipolar Couplings and Small- and Wide-Angle X-Ray Scattering.
J.Am.Chem.Soc., 132, 2010
|
|
1CXW
 
 | | THE SECOND TYPE II MODULE FROM HUMAN MATRIX METALLOPROTEINASE 2 | | 分子名称: | HUMAN MATRIX METALLOPROTEINASE 2 | | 著者 | Briknarova, K, Grishaev, A, Banyai, L, Tordai, H, Patthy, L, Llinas, M. | | 登録日 | 1999-08-31 | | 公開日 | 1999-11-12 | | 最終更新日 | 2022-12-21 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The second type II module from human matrix metalloproteinase 2: structure, function and dynamics.
Structure Fold.Des., 7, 1999
|
|
2MJB
 
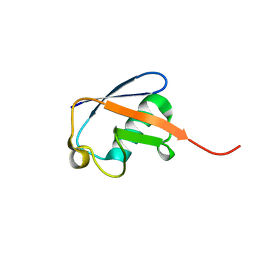 | | Solution nmr structure of ubiquitin refined against dipolar couplings in 4 media | | 分子名称: | Ubiquitin-60S ribosomal protein L40 | | 著者 | Maltsev, A, Grishaev, A, Roche, J, Zasloff, M, Bax, A. | | 登録日 | 2014-01-02 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Improved cross validation of a static ubiquitin structure derived from high precision residual dipolar couplings measured in a drug-based liquid crystalline phase.
J.Am.Chem.Soc., 136, 2014
|
|
2JWL
 
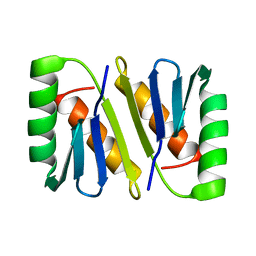 | |
2MK3
 
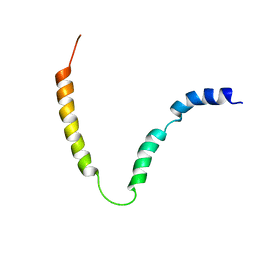 | | Solution NMR structure of gp41 ectodomain monomer on a DPC micelle | | 分子名称: | Transmembrane glycoprotein, chimeric construct | | 著者 | Roche, J, Louis, J.M, Grishaev, A, Ying, J, Bax, A. | | 登録日 | 2014-01-23 | | 公開日 | 2014-02-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Dissociation of the trimeric gp41 ectodomain at the lipid-water interface suggests an active role in HIV-1 Env-mediated membrane fusion.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2KX9
 
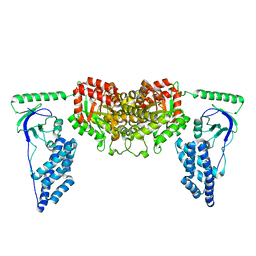 | | Solution Structure of the Enzyme I dimer Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | 分子名称: | Phosphoenolpyruvate-protein phosphotransferase | | 著者 | Schwieters, C.D, Suh, J, Grishaev, A, Takayama, Y, Guirlando, R, Clore, G. | | 登録日 | 2010-04-29 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Solution structure of the 128 kDa enzyme I dimer from Escherichia coli and its 146 kDa complex with HPr using residual dipolar couplings and small- and wide-angle X-ray scattering.
J.Am.Chem.Soc., 132, 2010
|
|
2M8L
 
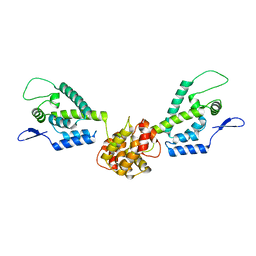 | | HIV capsid dimer structure | | 分子名称: | Capsid protein p24 | | 著者 | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | 登録日 | 2013-05-23 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2M8P
 
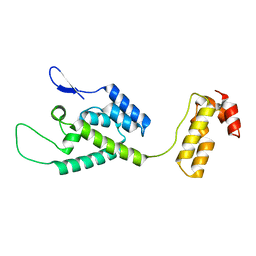 | | The structure of the W184AM185A mutant of the HIV-1 capsid protein | | 分子名称: | Capsid protein p24 | | 著者 | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | 登録日 | 2013-05-24 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2M8N
 
 | | HIV-1 capsid monomer structure | | 分子名称: | Capsid protein p24 | | 著者 | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | 登録日 | 2013-05-24 | | 公開日 | 2013-11-20 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2L5H
 
 | | Solution Structure of the H189Q mutant of the Enzyme I dimer Using Residual Dipolar Couplings and Small Angle X-Ray Scattering | | 分子名称: | Phosphoenolpyruvate-protein phosphotransferase | | 著者 | Takayama, Y.D, Schwieters, C.D, Grishaev, A, Guirlando, R, Clore, G. | | 登録日 | 2010-11-01 | | 公開日 | 2011-01-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Combined Use of Residual Dipolar Couplings and Solution X-ray Scattering To Rapidly Probe Rigid-Body Conformational Transitions in a Non-phosphorylatable Active-Site Mutant of the 128 kDa Enzyme I Dimer.
J.Am.Chem.Soc., 133, 2011
|
|
2N5T
 
 | | Ensemble solution structure of the phosphoenolpyruvate-Enzyme I complex from the bacterial phosphotransferase system | | 分子名称: | Phosphoenolpyruvate-protein phosphotransferase | | 著者 | Venditti, V, Schwieters, C.D, Grishaev, A, Clore, G. | | 登録日 | 2015-07-28 | | 公開日 | 2015-09-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Dynamic equilibrium between closed and partially closed states of the bacterial Enzyme I unveiled by solution NMR and X-ray scattering.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2KNF
 
 | | Solution structure and functional characterization of human plasminogen kringle 5 | | 分子名称: | Plasminogen | | 著者 | Battistel, M.D, Grishaev, A, An, S.A, Castellino, F.J, Llinas, M. | | 登録日 | 2009-08-21 | | 公開日 | 2009-10-27 | | 最終更新日 | 2021-10-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and functional characterization of human plasminogen kringle 5.
Biochemistry, 48, 2009
|
|
2LV6
 
 | | The complex between Ca-Calmodulin and skeletal muscle myosin light chain kinase from combination of NMR and aqueous and contrast-matched SAXS data | | 分子名称: | CALCIUM ION, Calmodulin, Myosin light chain kinase 2, ... | | 著者 | Grishaev, A.V, Anthis, N.J, Clore, G.M. | | 登録日 | 2012-06-29 | | 公開日 | 2013-02-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR, SOLUTION SCATTERING | | 主引用文献 | Contrast-matched small-angle X-ray scattering from a heavy-atom-labeled protein in structure determination: application to a lead-substituted calmodulin-peptide complex.
J.Am.Chem.Soc., 134, 2012
|
|
6B67
 
 | |
6UWT
 
 | |
6UWR
 
 | |
2OED
 
 | | GB3 solution structure obtained by refinement of X-ray structure with dipolar couplings | | 分子名称: | Immunoglobulin G-binding protein G | | 著者 | Ulmer, T.S, Ramirez, B.E, Delaglio, F, Bax, A, Grishaev, A. | | 登録日 | 2006-12-29 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Evaluation of backbone proton positions and dynamics in a small protein by liquid crystal NMR spectroscopy
J.Am.Chem.Soc., 125, 2003
|
|
6UWI
 
 | |
6UWO
 
 | |
