3FM9
 
 | | Analysis of the Structural Determinants Underlying Discrimination between Substrate and Solvent in beta-Phosphoglucomutase Catalysis | | 分子名称: | Beta-phosphoglucomutase, MAGNESIUM ION | | 著者 | Finci, L, Lahiri, S, Peisach, E, Allen, K.N. | | 登録日 | 2008-12-19 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Analysis of the structural determinants underlying discrimination between substrate and solvent in beta-phosphoglucomutase catalysis.
Biochemistry, 48, 2009
|
|
6STR
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N,N'-Diacetylchitobiose; 60 seconds soaking | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL | | 著者 | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | 登録日 | 2019-09-11 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STQ
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N,N'-Diacetylchitobiose; 30 seconds soaking | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL | | 著者 | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | 登録日 | 2019-09-11 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STN
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N-Acetyl glucosamine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL | | 著者 | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | 登録日 | 2019-09-11 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STM
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) | | 分子名称: | Arundo donax Lectin (ADL), GLYCEROL | | 著者 | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | 登録日 | 2019-09-11 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STP
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with sialic acid | | 分子名称: | Arundo donax Lectin (ADL), GLYCEROL, N-acetyl-alpha-neuraminic acid | | 著者 | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | 登録日 | 2019-09-11 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
6STO
 
 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N-Acetyl lactosamine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL, ... | | 著者 | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | 登録日 | 2019-09-11 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
1U0T
 
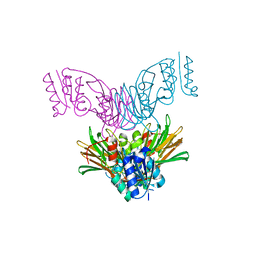 | | Crystal structure of Mycobacterium tuberculosis NAD kinase | | 分子名称: | Inorganic polyphosphate/ATP-NAD kinase | | 著者 | Garavaglia, S, Raffaelli, N, Finaurini, L, Magni, G, Rizzi, M, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2004-07-14 | | 公開日 | 2004-08-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A novel fold revealed by Mycobacterium tuberculosis NAD kinase, a key allosteric enzyme in NADP biosynthesis
J.Biol.Chem., 279, 2004
|
|
1U0R
 
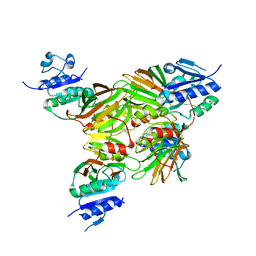 | | Crystal structure of Mycobacterium tuberculosis NAD kinase | | 分子名称: | Inorganic polyphosphate/ATP-NAD kinase | | 著者 | Garavaglia, S, Raffaelli, N, Finaurini, L, Magni, G, Rizzi, M, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2004-07-14 | | 公開日 | 2004-08-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A novel fold revealed by Mycobacterium tuberculosis NAD kinase, a key allosteric enzyme in NADP biosynthesis
J.Biol.Chem., 279, 2004
|
|
9FMD
 
 | |
1N49
 
 | | Viability of a Drug-Resistant HIV-1 Protease Variant: Structural Insights for Better Anti-Viral Therapy | | 分子名称: | Protease, RITONAVIR | | 著者 | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | 登録日 | 2002-10-30 | | 公開日 | 2003-01-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Viability of a Drug-Resistant Human Immunodeficiency Virus Type 1 Protease Variant:
Structural Insights for Better Antiviral Therapy
J.VIROL., 77, 2003
|
|
7ZNJ
 
 | | Structure of an ALYREF-exon junction complex hexamer | | 分子名称: | Eukaryotic initiation factor 4A-III, N-terminally processed, MAGNESIUM ION, ... | | 著者 | Pacheco-Fiallos, F.B, Vorlaender, M.K, Plaschka, C. | | 登録日 | 2022-04-21 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (2.4 Å) | | 主引用文献 | mRNA recognition and packaging by the human transcription-export complex.
Nature, 616, 2023
|
|
7ZNK
 
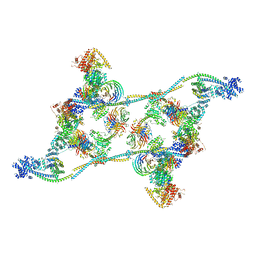 | | Structure of an endogenous human TREX complex bound to mRNA | | 分子名称: | RNA, Spliceosome RNA helicase DDX39B, THO complex subunit 1, ... | | 著者 | Pacheco-Fiallos, F.B, Vorlaender, M.K, Plaschka, C. | | 登録日 | 2022-04-21 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | mRNA recognition and packaging by the human transcription-export complex.
Nature, 616, 2023
|
|
7ZNL
 
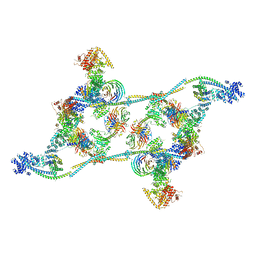 | | Structure of the human TREX core THO-UAP56 complex | | 分子名称: | Spliceosome RNA helicase DDX39B, THO complex subunit 1, THO complex subunit 2, ... | | 著者 | Pacheco-Fiallos, F.B, Vorlaender, M.K, Plaschka, C. | | 登録日 | 2022-04-21 | | 公開日 | 2023-05-17 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | mRNA recognition and packaging by the human transcription-export complex.
Nature, 616, 2023
|
|
7APK
 
 | | Structure of the human THO - UAP56 complex | | 分子名称: | Spliceosome RNA helicase DDX39B, THO complex subunit 1, THO complex subunit 2, ... | | 著者 | Hohmann, U, Puehringer, T, Plaschka, C. | | 登録日 | 2020-10-17 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of the human core transcription-export complex reveals a hub for multivalent interactions.
Elife, 9, 2020
|
|
8Q7V
 
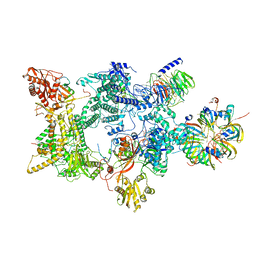 | | Structure of the recycling U5 snRNP bound to chaperones CD2BP2 and TSSC4 (State 1) | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | 登録日 | 2023-08-17 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7Q
 
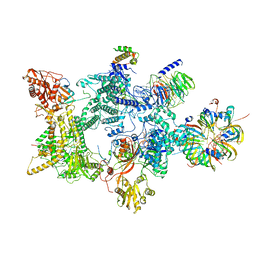 | | Structure of the recycling U5 snRNP bound to chaperones CD2BP2 and TSSC4 (State 2) | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | 登録日 | 2023-08-16 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7X
 
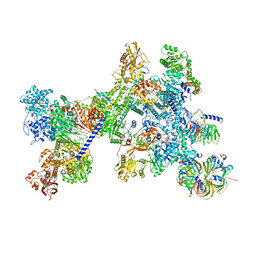 | | Structure of the recycling U5 snRNP bound to chaperone CD2BP2 (State 4) | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | 登録日 | 2023-08-17 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q7W
 
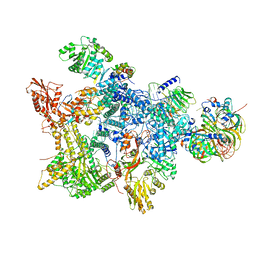 | | Structure of the recycling U5 snRNP bound to chaperone CD2BP2 (State 3) | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Riabov Bassat, D, Plaschka, C, Vorlaender, M.K. | | 登録日 | 2023-08-17 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis of human U5 snRNP late biogenesis and recycling.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1HXW
 
 | |
6WXW
 
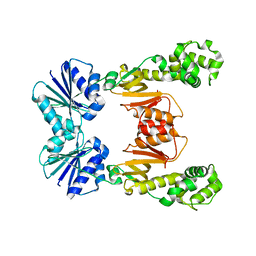 | |
6WXY
 
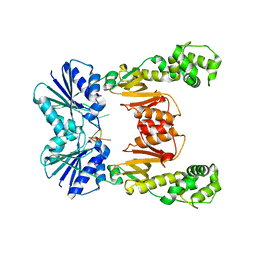 | | crystal structure of cA6-bound Card1 | | 分子名称: | Card1, cA6 | | 著者 | Rostol, J, Xie, W, Patel, D.J, Marraffini, L. | | 登録日 | 2020-05-12 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Card1 nuclease provides defence during type III CRISPR immunity.
Nature, 590, 2021
|
|
6WXX
 
 | | crystal structure of cA4-activated Card1 | | 分子名称: | Card1, MANGANESE (II) ION, cA4 | | 著者 | Rostol, J, Xie, W, Patel, D.J, Marraffini, L. | | 登録日 | 2020-05-12 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The Card1 nuclease provides defence during type III CRISPR immunity.
Nature, 590, 2021
|
|
6XL1
 
 | | crystal structure of cA4-activated Card1(D294N) | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Card1, MANGANESE (II) ION, ... | | 著者 | Rostol, J, Xie, W, Patel, D.J, Marraffini, L. | | 登録日 | 2020-06-27 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The Card1 nuclease provides defence during type III CRISPR immunity.
Nature, 590, 2021
|
|
1PSU
 
 | | Structure of the E. coli PaaI protein from the phyenylacetic acid degradation operon | | 分子名称: | Phenylacetic acid degradation protein PaaI | | 著者 | Kniewel, R, Buglino, J, Solorzano, V, Wu, J, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-06-21 | | 公開日 | 2003-07-08 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI.
J.Biol.Chem., 281, 2006
|
|
