1RN7
 
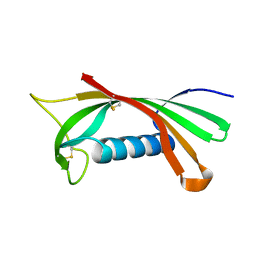 | | Structure of human cystatin D | | 分子名称: | Cystatin D | | 著者 | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | 登録日 | 2003-11-30 | | 公開日 | 2004-05-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
1ROA
 
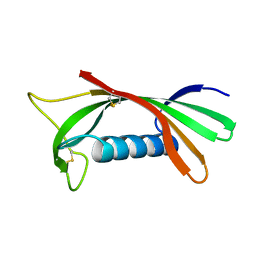 | | Structure of human cystatin D | | 分子名称: | Cystatin D | | 著者 | Alvarez-Fernandez, M, Liang, Y.H, Abrahamson, M, Su, X.D. | | 登録日 | 2003-12-01 | | 公開日 | 2004-05-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of human cystatin D, a cysteine peptidase inhibitor with restricted inhibition profile.
J.Biol.Chem., 280, 2005
|
|
8BMV
 
 | | Ligand binding domain of the P. Putida receptor McpH in complex with Uric acid | | 分子名称: | Methyl-accepting chemotaxis protein McpH, URIC ACID | | 著者 | Gavira, J.A, Krell, T, Fernandez, M, Martinez-Rodriguez, S. | | 登録日 | 2022-11-11 | | 公開日 | 2024-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Ubiquitous purine sensor modulates diverse signal transduction pathways in bacteria.
Nat Commun, 15, 2024
|
|
6S37
 
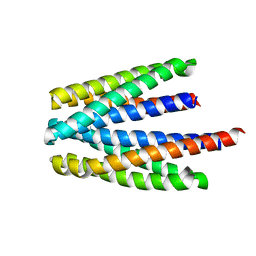 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with salicylic acid | | 分子名称: | 2-HYDROXYBENZOIC ACID, ACETATE ION, Aromatic acid chemoreceptor | | 著者 | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | 登録日 | 2019-06-24 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S18
 
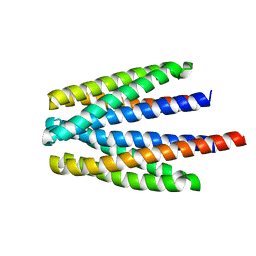 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with glycerol | | 分子名称: | Aromatic acid chemoreceptor, CHLORIDE ION, GLYCEROL | | 著者 | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S1A
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP | | 分子名称: | Aromatic acid chemoreceptor, SULFATE ION | | 著者 | Gavira, J.A, Matilla, M.A, Fernandez, M, Krell, T. | | 登録日 | 2019-06-18 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.112 Å) | | 主引用文献 | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S3B
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with benzoate | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Aromatic acid chemoreceptor, ... | | 著者 | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | 登録日 | 2019-06-24 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S38
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with quinate | | 分子名称: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, Aromatic acid chemoreceptor | | 著者 | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | 登録日 | 2019-06-24 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
6S33
 
 | | Ligand binding domain of the P. putida receptor PcaY_PP in complex with Protocatechuate | | 分子名称: | 3,4-DIHYDROXYBENZOIC ACID, ACETATE ION, Aromatic acid chemoreceptor | | 著者 | Gavira, J.A, Mantilla, M.A, Fernandez, M, Krell, T. | | 登録日 | 2019-06-24 | | 公開日 | 2020-10-21 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | The structural basis for signal promiscuity in a bacterial chemoreceptor.
Febs J., 288, 2021
|
|
3G73
 
 | | Structure of the FOXM1 DNA binding | | 分子名称: | DNA (5'-D(P*AP*AP*AP*TP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*GP*CP*CP*CP*G)-3'), DNA (5'-D(P*TP*TP*CP*GP*GP*GP*CP*TP*GP*TP*TP*TP*AP*TP*AP*AP*AP*CP*AP*AP*T)-3'), Forkhead box protein M1, ... | | 著者 | Littler, D.R, Perrakis, A, Hibbert, R.G, Medema, R.H. | | 登録日 | 2009-02-09 | | 公開日 | 2009-03-03 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence
Nucleic Acids Res., 2010
|
|
3QVM
 
 | | The structure of olei00960, a hydrolase from Oleispira antarctica | | 分子名称: | CALCIUM ION, CHLORIDE ION, Olei00960, ... | | 著者 | Singer, A.U, Kagan, O, Kim, Y, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-02-25 | | 公開日 | 2011-04-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
7S0E
 
 | |
7S0D
 
 | |
7S0B
 
 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody N-612-056 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, N-612-056 Fab Heavy Chain, N-612-056 Light Chain, ... | | 著者 | Tanaka, S, Barnes, C.O, Bjorkman, P.J. | | 登録日 | 2021-08-30 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Rapid identification of neutralizing antibodies against SARS-CoV-2 variants by mRNA display.
Cell Rep, 38, 2022
|
|
7S0C
 
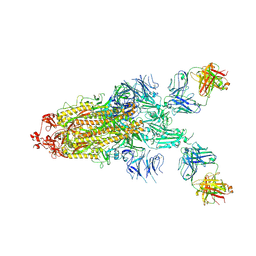 | |
2NQD
 
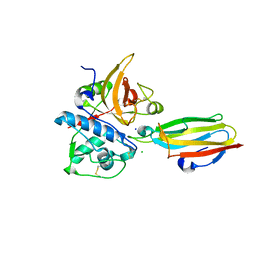 | | Crystal structure of cysteine protease inhibitor, chagasin, in complex with human cathepsin L | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Cathepsin L, ... | | 著者 | Redzynia, I, Bujacz, G, Ljunggren, A, Jaskolski, M, Abrahamson, M. | | 登録日 | 2006-10-31 | | 公開日 | 2007-07-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease
J.Mol.Biol., 371, 2007
|
|
2NNR
 
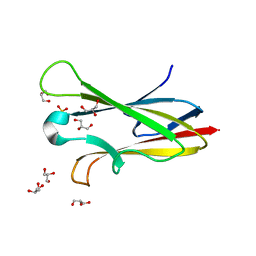 | | Crystal structure of chagasin, cysteine protease inhibitor from Trypanosoma cruzi | | 分子名称: | CHLORIDE ION, Chagasin, GLYCEROL, ... | | 著者 | Redzynia, I, Bujacz, G, Ljunggren, A, Jaskolski, M, Abrahamson, M. | | 登録日 | 2006-10-24 | | 公開日 | 2007-07-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the parasite protease inhibitor chagasin in complex with a host target cysteine protease
J.Mol.Biol., 371, 2007
|
|
3LMB
 
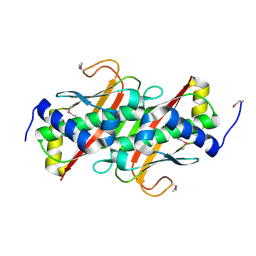 | | The crystal structure of the protein OLEI01261 with unknown function from Chlorobaculum tepidum TLS | | 分子名称: | Uncharacterized protein | | 著者 | Zhang, R, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-01-29 | | 公開日 | 2010-03-16 | | 最終更新日 | 2013-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3LNP
 
 | | Crystal Structure of Amidohydrolase family Protein OLEI01672_1_465 from Oleispira antarctica | | 分子名称: | ACETIC ACID, Amidohydrolase family Protein OLEI01672_1_465, CALCIUM ION, ... | | 著者 | Kim, Y, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-02-02 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3M16
 
 | | Structure of a Transaldolase from Oleispira antarctica | | 分子名称: | Transaldolase | | 著者 | Singer, A.U, Kagan, O, Zhang, R, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-03-04 | | 公開日 | 2010-06-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
2YJN
 
 | | Structure of the glycosyltransferase EryCIII from the erythromycin biosynthetic pathway, in complex with its activating partner, EryCII | | 分子名称: | DTDP-4-KETO-6-DEOXY-HEXOSE 3,4-ISOMERASE, GLYCOSYLTRANSFERASE | | 著者 | Moncrieffe, M.C, Fernandez, M.J, Spiteller, D, Matsumura, H, Gay, N.J, Luisi, B.F, Leadlay, P.F. | | 登録日 | 2011-05-20 | | 公開日 | 2011-11-09 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.091 Å) | | 主引用文献 | Structure of the Glycosyltransferase Eryciii in Complex with its Activating P450 Homologue Erycii.
J.Mol.Biol., 415, 2012
|
|
3LQY
 
 | | Crystal structure of putative isochorismatase hydrolase from Oleispira antarctica | | 分子名称: | GLYCEROL, putative isochorismatase hydrolase | | 著者 | Goral, A, Chruszcz, M, Kagan, O, Cymborowski, M, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-02-10 | | 公開日 | 2010-03-16 | | 最終更新日 | 2022-04-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of a putative isochorismatase hydrolase from Oleispira antarctica.
J.Struct.Funct.Genom., 13, 2012
|
|
3VCR
 
 | | Crystal structure of a putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase from Oleispira antarctica | | 分子名称: | PYRUVIC ACID, putative Kdpg (2-keto-3-deoxy-6-phosphogluconate) aldolase | | 著者 | Stogios, P.J, Kagan, O, Di Leo, R, Yim, V, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-01-04 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3V77
 
 | | Crystal structure of a putative fumarylacetoacetate isomerase/hydrolase from Oleispira antarctica | | 分子名称: | ACETATE ION, D(-)-TARTARIC ACID, Putative fumarylacetoacetate isomerase/hydrolase, ... | | 著者 | Stogios, P.J, Kagan, O, Di Leo, R, Bochkarev, A, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-12-20 | | 公開日 | 2012-01-18 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
1TIJ
 
 | | 3D Domain-swapped human cystatin C with amyloid-like intermolecular beta-sheets | | 分子名称: | Cystatin C | | 著者 | Janowski, R, Kozak, M, Abrahamson, M, Grubb, A, Jaskolski, M. | | 登録日 | 2004-06-02 | | 公開日 | 2005-07-19 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.03 Å) | | 主引用文献 | 3D domain-swapped human cystatin C with amyloidlike intermolecular beta-sheets.
Proteins, 61, 2005
|
|
