5V6C
 
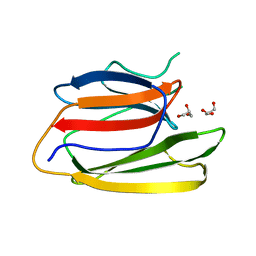 | | Crystal Structure of the Second beta-Prism Domain of RbmC from V. cholerae | | 分子名称: | GLYCEROL, Hemolysin-related protein | | 著者 | De, S, Kaus, K, Sinclair, S, Case, B.C, Olson, R. | | 登録日 | 2017-03-16 | | 公開日 | 2018-01-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of mammalian glycan targeting by Vibrio cholerae cytolysin and biofilm proteins.
PLoS Pathog., 14, 2018
|
|
3O44
 
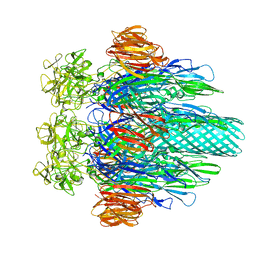 | |
2MD5
 
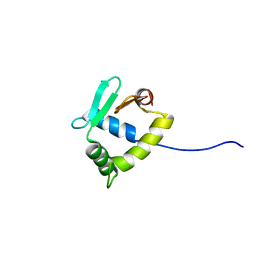 | | Structure of uninhibited ETV6 ETS domain | | 分子名称: | Transcription factor ETV6 | | 著者 | De, S, Mcintosh, L.P, Chan, A.C, Coyne, H.J, Okon, M, Graves, B.J, Murphy, M.E. | | 登録日 | 2013-08-29 | | 公開日 | 2013-12-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
5V6F
 
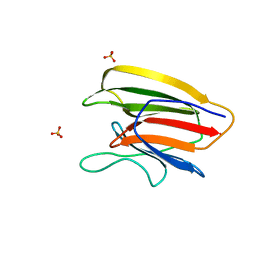 | | Crystal Structure of the Second beta-Prism Domain of RbmC from V. cholerae bound to Mannotriose | | 分子名称: | Hemolysin-related protein, SULFATE ION, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | 著者 | De, S, Kaus, K, Sinclair, S, Case, B.C, Olson, R. | | 登録日 | 2017-03-16 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis of mammalian glycan targeting by Vibrio cholerae cytolysin and biofilm proteins.
PLoS Pathog., 14, 2018
|
|
5V6K
 
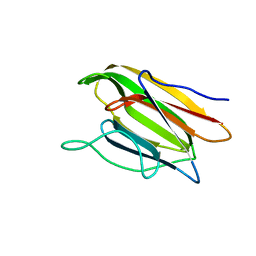 | | Crystal Structure of the Second beta-Prism Domain of RbmC from V. cholerae Bound to N-acetylglucosaminyl-beta-1,2-mannose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, GLYCEROL, Hemolysin-related protein | | 著者 | De, S, Kaus, K, Sinclair, S, Case, B.C, Olson, R. | | 登録日 | 2017-03-16 | | 公開日 | 2018-01-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of mammalian glycan targeting by Vibrio cholerae cytolysin and biofilm proteins.
PLoS Pathog., 14, 2018
|
|
5D13
 
 | |
4MHG
 
 | | Crystal structure of ETV6 bound to a specific DNA sequence | | 分子名称: | Complementary Specific 14 bp DNA, Specific 14 bp DNA, Transcription factor ETV6 | | 著者 | Chan, A.C, De, S, Coyne III, H.J, Okon, M, Murphy, M.E, Graves, B.J, McIntosh, L.P. | | 登録日 | 2013-08-29 | | 公開日 | 2014-01-08 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Steric Mechanism of Auto-Inhibitory Regulation of Specific and Non-Specific DNA Binding by the ETS Transcriptional Repressor ETV6.
J.Mol.Biol., 426, 2014
|
|
7SYN
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 8(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYJ
 
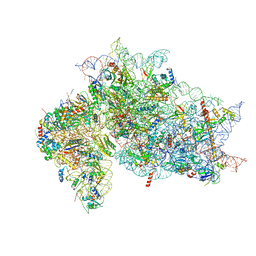 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 4(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYL
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, closed conformation. Structure 6(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYX
 
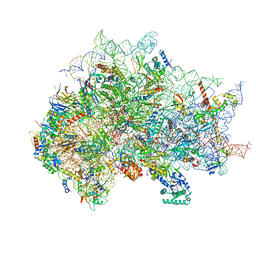 | | Structure of the delta dII IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S24, 40S ribosomal protein S25, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYW
 
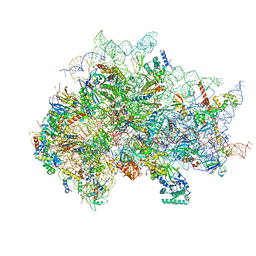 | | Structure of the wt IRES eIF5B-containing 48S initiation complex, closed conformation. Structure 15(wt) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-02-01 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYG
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 1(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
7SYI
 
 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 3(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYM
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head opening. Structure 7(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, 40S ribosomal protein S21, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Comprehensive structural overview of the HCV IRES-mediated translation initiation pathway
To Be Published
|
|
7EB1
 
 | | Solution NMR structure of the RRM domain of RNA binding protein RBM3 from homo sapiens | | 分子名称: | RNA-binding protein 3 | | 著者 | Boral, S, Roy, S, Basak, A.J, Maiti, S, Lee, W, De, S. | | 登録日 | 2021-03-08 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and dynamic studies of the human RNA binding protein RBM3 reveals the molecular basis of its oligomerization and RNA recognition.
Febs J., 289, 2022
|
|
7CFV
 
 | |
6UU8
 
 | | E. coli mutant sigma-S transcription initiation complex with a 7-nt RNA ("Fresh" mutant crystal soaked with GTP, UTP, and CTP for 30 minutes) | | 分子名称: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Zuo, Y, De, S, Steitz, T.A. | | 登録日 | 2019-10-30 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (4.4 Å) | | 主引用文献 | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UTW
 
 | | E. coli sigma-S transcription initiation complex with a 4-nt RNA ("Fresh" crystal) | | 分子名称: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Zuo, Y, De, S, Steitz, T.A. | | 登録日 | 2019-10-30 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.854 Å) | | 主引用文献 | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU0
 
 | | E. coli sigma-S transcription initiation complex with a 3-nt RNA and a mismatching GTP ("Fresh" crystal soaked with GTP for 1 hour) | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Zuo, Y, De, S, Steitz, T.A. | | 登録日 | 2019-10-30 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU6
 
 | | E. coli sigma-S transcription initiation complex with a 4-nt RNA and a UTP ("Old" crystal soaked with UTP, ddCTP, and dinucleotide ApG for 30 minutes) | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Zuo, Y, De, S, Steitz, T.A. | | 登録日 | 2019-10-30 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (4.201 Å) | | 主引用文献 | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UU1
 
 | | E. coli sigma-S transcription initiation complex with a 4-nt RNA and a CTP ("Fresh" crystal soaked with CTP, GTP, and ddTTP for 30 minutes) | | 分子名称: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Zuo, Y, De, S, Steitz, T.A. | | 登録日 | 2019-10-30 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (4.097 Å) | | 主引用文献 | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UTZ
 
 | | E. coli sigma-S transcription initiation complex with a 6-nt RNA ("Fresh" crystal soaked with CTP and UTP for 30 minutes) | | 分子名称: | DIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Zuo, Y, De, S, Steitz, T.A. | | 登録日 | 2019-10-30 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.803 Å) | | 主引用文献 | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UTY
 
 | | E. coli sigma-S transcription initiation complex with a mismatching CTP ("Old" crystal soaked with CTP for 30 minutes) | | 分子名称: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Zuo, Y, De, S, Steitz, T.A. | | 登録日 | 2019-10-30 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (4.15 Å) | | 主引用文献 | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
6UTX
 
 | | E. coli sigma-S transcription initiation complex with an empty bubble ("Old" crystal) | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Zuo, Y, De, S, Steitz, T.A. | | 登録日 | 2019-10-30 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (4.05 Å) | | 主引用文献 | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
