6GJ4
 
 | | Tubulin-6j complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-(quinolin-5-yl)naphtho[2,3-b]pyrrolo[1,2-d][1,4]oxazepin-4-yl acetate, CALCIUM ION, ... | | 著者 | Brindisi, M, Ulivieri, C, Alfano, G, Gemma, S, Balaguer, F.d.A, Khan, T, Grillo, A, Chemi, G, Menchon, G, Prota, A.E, Olieric, N, Agell, D.L, Barasoain, I, Diaz, J.F, Nebbioso, A, Conte, M.R, Lopresti, L, Magnano, S, Amet, R, Kinsella, P, Zisterer, D.M, Ibrahim, O, O'Sullivan, J, Morbidelli, L, Spaccapelo, R, Baldari, C, Butini, S, Novellino, E, Campiani, G, Altucci, L, Steinmetz, M.O, Brogi, S. | | 登録日 | 2018-05-16 | | 公開日 | 2018-12-05 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-activity relationships, biological evaluation and structural studies of novel pyrrolonaphthoxazepines as antitumor agents.
Eur J Med Chem, 162, 2018
|
|
4K1F
 
 | | Crystal structure of reduced tryparedoxin peroxidase from leishmania major at 2.34 A resolution | | 分子名称: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Ilari, A, Fiorillo, A, Di Chiaro, F. | | 登録日 | 2013-04-05 | | 公開日 | 2014-04-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structure-based discovery of the first non-covalent inhibitors of Leishmania major tryparedoxin peroxidase by high throughput docking
Sci Rep, 5, 2015
|
|
5WLO
 
 | | a novel 13-ring macrocyclic HIV-1 protease inhibitors involving the P1'-P2' ligands | | 分子名称: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-[(7E)-13-methoxy-1,1-dioxo-1,4,5,6,9,11-hexahydro-10,1lambda~6~,2-benzoxathiazacyclotridecin-2(3H)-yl]-1-phenylbutan-2-yl}carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2017-07-27 | | 公開日 | 2017-10-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Design, synthesis, X-ray studies, and biological evaluation of novel macrocyclic HIV-1 protease inhibitors involving the P1'-P2' ligands.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6V7A
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with NF2657 | | 分子名称: | Hdac6 protein, N-hydroxy-4-[(1-methyl-1,2-dihydro-1'H-spiro[indole-3,4'-piperidin]-1'-yl)methyl]benzamide, POTASSIUM ION, ... | | 著者 | Osko, J.D, Christianson, D.W. | | 登録日 | 2019-12-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.0874176 Å) | | 主引用文献 | Spiroindoline-Capped Selective HDAC6 Inhibitors: Design, Synthesis, Structural Analysis, and Biological Evaluation.
Acs Med.Chem.Lett., 11, 2020
|
|
6V79
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with NF2376 | | 分子名称: | 1,2-ETHANEDIOL, 4-{[(2S)-3,3-dimethyl-2-(pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]methyl}-N-hydroxybenzamide, Hdac6 protein, ... | | 著者 | Osko, J.D, Christianson, D.W. | | 登録日 | 2019-12-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.03951526 Å) | | 主引用文献 | Harnessing the Role of HDAC6 in Idiopathic Pulmonary Fibrosis: Design, Synthesis, Structural Analysis, and Biological Evaluation of Potent Inhibitors.
J.Med.Chem., 64, 2021
|
|
7Q5F
 
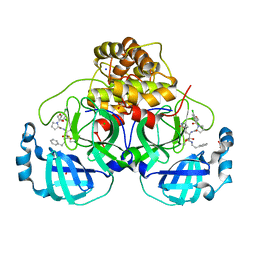 | | Crystal structure of F2F-2020216-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (S)-1-(2-(2,4-dichlorophenoxy)acetyl)-N-((S)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)-4-(phenethylamino)butan-2-yl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 3C-like proteinase, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2021-11-03 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Easy access to alpha-ketoamides as SARS-CoV-2 and MERS M pro inhibitors via the PADAM oxidation route.
Eur.J.Med.Chem., 244, 2022
|
|
7Q5E
 
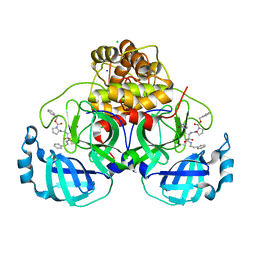 | | Crystal structure of F2F-2020209-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | 3C-like proteinase, CHLORIDE ION, SODIUM ION, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2021-11-03 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Easy access to alpha-ketoamides as SARS-CoV-2 and MERS M pro inhibitors via the PADAM oxidation route.
Eur.J.Med.Chem., 244, 2022
|
|
3S2V
 
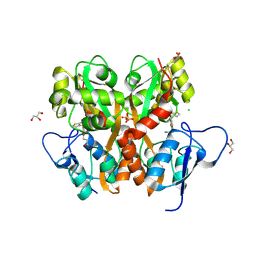 | | Crystal Structure of the Ligand Binding Domain of GluK1 in Complex with an Antagonist (S)-1-(2'-Amino-2'-carboxyethyl)-3-[(2-carboxythien-3-yl)methyl]thieno[3,4-d]pyrimidin-2,4-dione at 2.5 A Resolution | | 分子名称: | (S)-1-(2'-AMINO-2'-CARBOXYETHYL)-3-[(2-CARBOXYTHIEN-3-YL)METHYL]THIENO[3,4-D]PYRIMIDIN-2,4-DIONE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2011-05-17 | | 公開日 | 2011-06-22 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Selective kainate receptor (GluK1) ligands structurally based upon 1H-cyclopentapyrimidin-2,4(1H,3H)-dione: synthesis, molecular modeling, and pharmacological and biostructural characterization.
J.Med.Chem., 54, 2011
|
|
5UOV
 
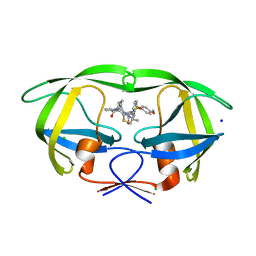 | | HIV-1 wild Type protease with GRL-1118A , an isophthalamide-derived P2-P3 ligand with the sulfonamide isostere as the P2' group | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2017-02-01 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Design of novel HIV-1 protease inhibitors incorporating isophthalamide-derived P2-P3 ligands: Synthesis, biological evaluation and X-ray structural studies of inhibitor-HIV-1 protease complex.
Bioorg. Med. Chem., 25, 2017
|
|
5UPZ
 
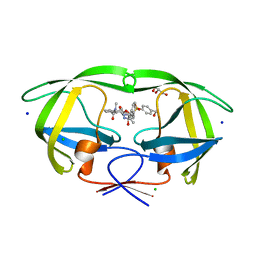 | | HIV-1 wild Type protease with GRL-0518A , an isophthalamide-derived P2-P3 ligand with the para-hydoxymethyl sulfonamide isostere as the P2' group | | 分子名称: | CHLORIDE ION, GLYCEROL, N~3~-{(2S,3R)-3-hydroxy-4-[{[4-(hydroxymethyl)phenyl]sulfonyl}(2-methylpropyl)amino]-1-phenylbutan-2-yl}-N~1~-methyl-N~1~-[(4-methyl-1,3-oxazol-2-yl)methyl]benzene-1,3-dicarboxamide, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2017-02-05 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Design of novel HIV-1 protease inhibitors incorporating isophthalamide-derived P2-P3 ligands: Synthesis, biological evaluation and X-ray structural studies of inhibitor-HIV-1 protease complex.
Bioorg. Med. Chem., 25, 2017
|
|
6B4N
 
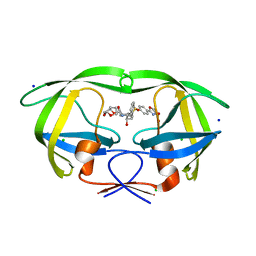 | | a hydroxymethyl functionality at the 4-position of the 2-phenyloxazole moiety of HIV-1 protease inhibitors involving the P2' ligands | | 分子名称: | CHLORIDE ION, Protease, SODIUM ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2017-09-27 | | 公開日 | 2017-11-22 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Design, Synthesis, Biological Evaluation, and X-ray Studies of HIV-1 Protease Inhibitors with Modified P2' Ligands of Darunavir.
ChemMedChem, 12, 2017
|
|
6F29
 
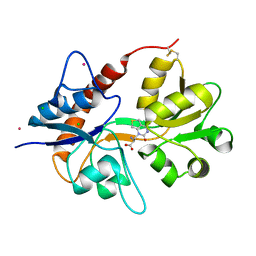 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-1-[2-Amino-2-carboxyethyl]-5,7-dihydrothieno[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 2.6A | | 分子名称: | (2~{S})-2-azanyl-3-[2,4-bis(oxidanylidene)-5,7-dihydrothieno[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2017-11-23 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | ( S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic Acid (AMPA) and Kainate Receptor Ligands: Further Exploration of Bioisosteric Replacements and Structural and Biological Investigation.
J. Med. Chem., 61, 2018
|
|
6CDJ
 
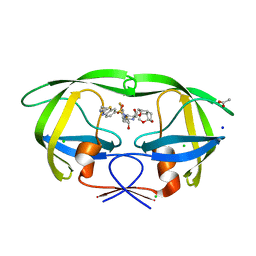 | | HIV-1 wild type protease with GRL-03314A, 6-5-5-ring fused umbrella-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | 分子名称: | (2aS,4R,4aS,7aS,7bS)-octahydro-2H-1,7-dioxacyclopenta[cd]inden-4-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2018-02-08 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Design and Synthesis of Highly Potent HIV-1 Protease Inhibitors Containing Tricyclic Fused Ring Systems as Novel P2 Ligands: Structure-Activity Studies, Biological and X-ray Structural Analysis.
J. Med. Chem., 61, 2018
|
|
6F28
 
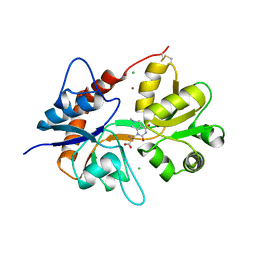 | | Crystal structure of the kainate receptor GluK3 ligand binding domain in complex with (S)-1-[2'-Amino-2'-carboxyethyl]-6-methyl-5,7-dihydropyrrolo[3,4-d]pyrimidin-2,4(1H,3H)-dione at resolution 2.4A | | 分子名称: | (2~{S})-2-azanyl-3-[6-methyl-2,4-bis(oxidanylidene)-5,7-dihydropyrrolo[3,4-d]pyrimidin-1-yl]propanoic acid, CHLORIDE ION, Glutamate receptor ionotropic, ... | | 著者 | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2017-11-23 | | 公開日 | 2018-02-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | ( S)-2-Amino-3-(5-methyl-3-hydroxyisoxazol-4-yl)propanoic Acid (AMPA) and Kainate Receptor Ligands: Further Exploration of Bioisosteric Replacements and Structural and Biological Investigation.
J. Med. Chem., 61, 2018
|
|
6CDL
 
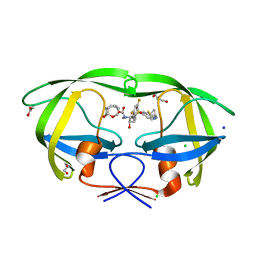 | | HIV-1 wild type protease with GRL-03214A, 6-5-5-ring fused umbrella-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | 分子名称: | (2aR,4S,4aR,7aR,7bR)-octahydro-2H-1,7-dioxacyclopenta[cd]inden-4-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2018-02-08 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Design and Synthesis of Highly Potent HIV-1 Protease Inhibitors Containing Tricyclic Fused Ring Systems as Novel P2 Ligands: Structure-Activity Studies, Biological and X-ray Structural Analysis.
J. Med. Chem., 61, 2018
|
|
6BZ2
 
 | | Crystal structure of wild-type HIV-1 protease with a novel HIV-1 inhibitor GRL-14213A of 6-5-5-ring fused crown-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | 著者 | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | 登録日 | 2017-12-22 | | 公開日 | 2018-02-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Design of Highly Potent, Dual-Acting and Central-Nervous-System-Penetrating HIV-1 Protease Inhibitors with Excellent Potency against Multidrug-Resistant HIV-1 Variants.
ChemMedChem, 13, 2018
|
|
5V0N
 
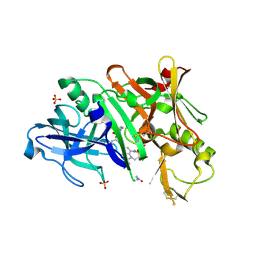 | | BACE1 in complex with inhibitor 5g | | 分子名称: | Beta-secretase 1, GLYCEROL, N-{(1S,2S)-1-[(2S)-4-benzyl-3-oxopiperazin-2-yl]-1-hydroxy-3-phenylpropan-2-yl}-7-ethyl-1,3,3-trimethyl-2,2-dioxo-1,2,3,4-tetrahydro-2lambda~6~-[1,2,5]thiadiazepino[3,4,5-hi]indole-9-carboxamide, ... | | 著者 | Mesecar, A, Ghosh, A, Yen, Y.-C. | | 登録日 | 2017-02-28 | | 公開日 | 2017-05-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.155 Å) | | 主引用文献 | Design, synthesis, and X-ray structural studies of BACE-1 inhibitors containing substituted 2-oxopiperazines as P1'-P2' ligands.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8OKL
 
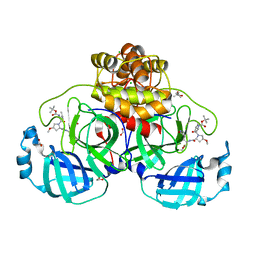 | | Crystal structure of F2F-2020185-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKM
 
 | | Crystal structure of F2F-2020197-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKK
 
 | | Crystal structure of F2F-2020184-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKN
 
 | | Crystal structure of F2F-2020198-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | 著者 | Costanzi, E, Demitri, N, Storici, P. | | 登録日 | 2023-03-28 | | 公開日 | 2023-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
6DHC
 
 | | X-ray structure of BACE1 in complex with a bicyclic isoxazoline carboxamide as the P3 ligand | | 分子名称: | (3R,3aR,6aS)-N-[(4R,7S,8S,10R,13S)-8-hydroxy-10,17-dimethyl-7-(2-methylpropyl)-5,11,14-trioxo-13-(propan-2-yl)-2-thia-6,12,15-triazaoctadecan-4-yl]hexahydrofuro[3,2-d][1,2]oxazole-3-carboxamide, Beta-secretase 1, GLYCEROL, ... | | 著者 | Mesecar, A.D, Lendy, E.K. | | 登録日 | 2018-05-19 | | 公開日 | 2018-07-25 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Design, synthesis, X-ray studies, and biological evaluation of novel BACE1 inhibitors with bicyclic isoxazoline carboxamides as the P3 ligand.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
