4XXO
 
 | |
7KYD
 
 | | Drosophila melanogaster long-chain fatty-acyl-CoA synthetase CG6178 | | 分子名称: | 1,2-ETHANEDIOL, 5'-O-[(S)-hydroxy(octanoyloxy)phosphoryl]adenosine, Long-chain fatty-acyl-CoA synthetase CG6178 | | 著者 | Adams, S.T, Zephyr, J, Bohn, M.F, Schiffer, C.A, Miller, S.C. | | 登録日 | 2020-12-07 | | 公開日 | 2022-01-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | FruitFire: a luciferase based on a fruit fly metabolic enzyme.
Biorxiv, 2023
|
|
4IOU
 
 | |
7TCZ
 
 | | Human cytomegalovirus protease mutant (C84A, C87A, C138A, C202A) in complex with inhibitor | | 分子名称: | Assemblin, [1-(2-oxopropyl)-4-phenyl-1H-1,2,3-triazol-5-yl]methyl benzylcarbamate | | 著者 | Hulce, K.R, Bohn, M, Ongpipattanakul, C, Jaishankar, P, Renslo, A.R, Craik, C.S. | | 登録日 | 2021-12-29 | | 公開日 | 2022-01-12 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Inhibiting a dynamic viral protease by targeting a non-catalytic cysteine.
Cell Chem Biol, 29, 2022
|
|
6EFK
 
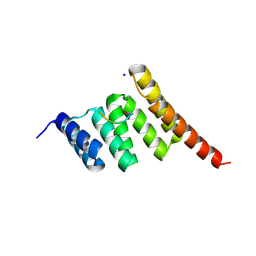 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated HSP70 peptide | | 分子名称: | ACE-ILE-GLU-GLU-VAL-ASP, E3 ubiquitin-protein ligase CHIP, SODIUM ION | | 著者 | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | 登録日 | 2018-08-16 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
6NSV
 
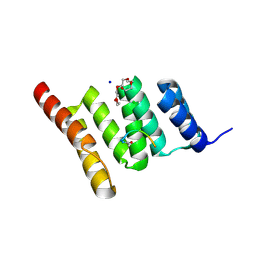 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated optimized peptide | | 分子名称: | ACE-LEU-TRP-TRP-PRO-ASP, CHLORIDE ION, E3 ubiquitin-protein ligase CHIP, ... | | 著者 | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | 登録日 | 2019-01-25 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.305 Å) | | 主引用文献 | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
7KKH
 
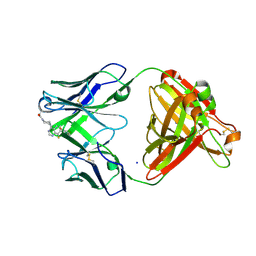 | | P1A4 Fab in complex with ARS1620 | | 分子名称: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, P1A4 Fab Heavy Chain, P1A4 Fab Light Chain, ... | | 著者 | Basu, K, Rohweder, P.J, Zhang, Z, Bohn, M.-F, Shokat, K, Craik, C.S. | | 登録日 | 2020-10-27 | | 公開日 | 2022-04-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A covalent inhibitor of K-Ras(G12C) induces MHC class I presentation of haptenated peptide neoepitopes targetable by immunotherapy.
Cancer Cell, 40, 2022
|
|
5KEG
 
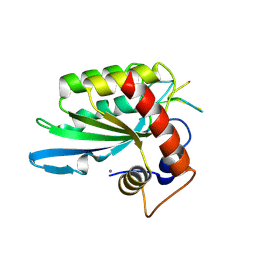 | | Crystal structure of APOBEC3A in complex with a single-stranded DNA | | 分子名称: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*TP*TP*CP*TP*T)-3'), ... | | 著者 | Kouno, T, Hilbert, B.J, Silvas, T, Royer, W.E, Matsuo, H, Schiffer, C.A. | | 登録日 | 2016-06-09 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of APOBEC3A bound to single-stranded DNA reveals structural basis for cytidine deamination and specificity.
Nat Commun, 8, 2017
|
|
8GCK
 
 | | Crystal structure of the human CHIP-TPR domain in complex with a 6mer acetylated tau peptide | | 分子名称: | ACE-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | 著者 | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | 登録日 | 2023-03-02 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.36823535 Å) | | 主引用文献 | Intersecting PTMs regulate clearance of pathogenic tau by the ubiquitin ligase CHIP.
To Be Published
|
|
5V5D
 
 | | Room temperature (280K) crystal structure of Kaposi's sarcoma-associated herpesvirus protease in complex with allosteric inhibitor (compound 250) | | 分子名称: | 4-{[6-(cyclohexylmethyl)pyridine-2-carbonyl]amino}-3-(phenylamino)benzoic acid, ORF 17 | | 著者 | Thompson, M.C, Acker, T.M, Fraser, J.S, Craik, C.S. | | 登録日 | 2017-03-14 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.104 Å) | | 主引用文献 | Allosteric Inhibitors, Crystallography, and Comparative Analysis Reveal Network of Coordinated Movement across Human Herpesvirus Proteases.
J. Am. Chem. Soc., 139, 2017
|
|
5V5E
 
 | | Room temperature (280K) crystal structure of Kaposi's sarcoma-associated herpesvirus protease in complex with allosteric inhibitor (compound 733) | | 分子名称: | 4-{[6-(cyclohexylmethyl)pyridine-2-carbonyl]amino}-3-{[3-(trifluoromethoxy)phenyl]amino}benzoic acid, ORF 17 | | 著者 | Thompson, M.C, Acker, T.M, Fraser, J.S, Craik, C.S. | | 登録日 | 2017-03-14 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.299 Å) | | 主引用文献 | Allosteric Inhibitors, Crystallography, and Comparative Analysis Reveal Network of Coordinated Movement across Human Herpesvirus Proteases.
J. Am. Chem. Soc., 139, 2017
|
|
8FYU
 
 | | Crystal structure of the human CHIP-TPR domain in complex with a 10mer acetylated tau peptide | | 分子名称: | ACE-SER-SER-THR-GLY-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | 著者 | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | 登録日 | 2023-01-26 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.84839141 Å) | | 主引用文献 | Phosphorylation of tau at a single residue inhibits binding to the E3 ubiquitin ligase, CHIP.
Nat Commun, 15, 2024
|
|
