6BTM
 
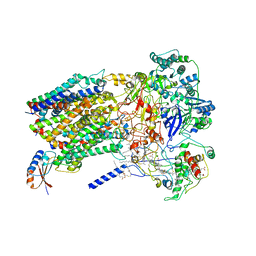 | | Structure of Alternative Complex III from Flavobacterium johnsoniae (Wild Type) | | 分子名称: | (2S)-3-hydroxypropane-1,2-diyl ditetradecanoate, Alternative Complex III subunit A, Alternative Complex III subunit B, ... | | 著者 | Sun, C, Benlekbir, S, Venkatakrishnan, P, Yuhang, W, Tajkhorshid, E, Rubinstein, J.L, Gennis, R.B. | | 登録日 | 2017-12-07 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure of the alternative complex III in a supercomplex with cytochrome oxidase.
Nature, 557, 2018
|
|
7TAP
 
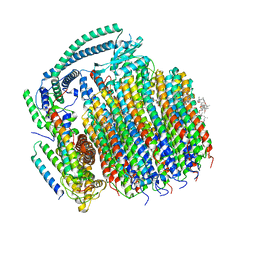 | | Cryo-EM structure of archazolid A bound to yeast VO V-ATPase | | 分子名称: | Archazolid A, V-type proton ATPase subunit a, vacuolar isoform, ... | | 著者 | Keon, K.A, Rubinstein, J.L, Benlekbir, S, Kirsch, S.H, Muller, R. | | 登録日 | 2021-12-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-EM of the Yeast V O Complex Reveals Distinct Binding Sites for Macrolide V-ATPase Inhibitors.
Acs Chem.Biol., 17, 2022
|
|
7TAO
 
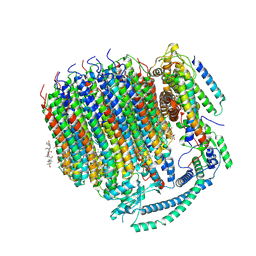 | | Cryo-EM structure of bafilomycin A1 bound to yeast VO V-ATPase | | 分子名称: | (5R)-2,4-dideoxy-1-C-{(2S,3R,4S)-3-hydroxy-4-[(2R,3S,4E,6E,9R,10S,11R,12E,14Z)-10-hydroxy-3,15-dimethoxy-7,9,11,13-tetramethyl-16-oxo-1-oxacyclohexadeca-4,6,12,14-tetraen-2-yl]pentan-2-yl}-4-methyl-5-propan-2-yl-alpha-D-threo-pentopyranose, V-type proton ATPase subunit a, vacuolar isoform, ... | | 著者 | Keon, K.A, Rubinstein, J.L, Benlekbir, S, Kirsch, S.H, Muller, R. | | 登録日 | 2021-12-21 | | 公開日 | 2022-02-23 | | 最終更新日 | 2022-03-30 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM of the Yeast V O Complex Reveals Distinct Binding Sites for Macrolide V-ATPase Inhibitors.
Acs Chem.Biol., 17, 2022
|
|
5KGF
 
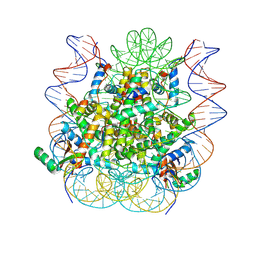 | | Structural model of 53BP1 bound to a ubiquitylated and methylated nucleosome, at 4.5 A resolution | | 分子名称: | DNA (145-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | 著者 | Wilson, M.D, Benlekbir, S, Sicheri, F, Rubinstein, J.L, Durocher, D. | | 登録日 | 2016-06-13 | | 公開日 | 2016-07-27 | | 最終更新日 | 2020-01-15 | | 実験手法 | ELECTRON MICROSCOPY (4.54 Å) | | 主引用文献 | The structural basis of modified nucleosome recognition by 53BP1.
Nature, 536, 2016
|
|
7MPE
 
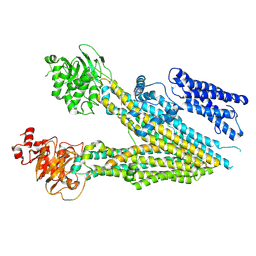 | |
6HWH
 
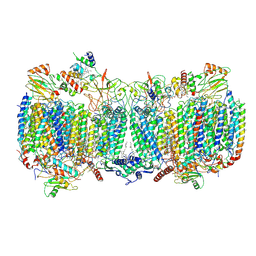 | | Structure of a functional obligate respiratory supercomplex from Mycobacterium smegmatis | | 分子名称: | CARDIOLIPIN, COPPER (II) ION, Co-purified unknown peptide built as polyALA, ... | | 著者 | Wiseman, B, Nitharwal, R.G, Fedotovskaya, O, Schafer, J, Guo, H, Kuang, Q, Benlekbir, S, Sjostrand, D, Adelroth, P, Rubinstein, J.L, Brzezinski, P, Hogbom, M. | | 登録日 | 2018-10-12 | | 公開日 | 2018-11-07 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of a functional obligate complex III2IV2respiratory supercomplex from Mycobacterium smegmatis.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5TJ5
 
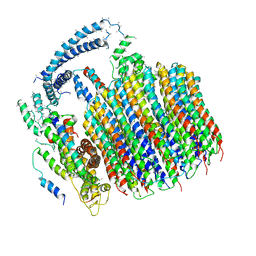 | | Atomic model for the membrane-embedded motor of a eukaryotic V-ATPase | | 分子名称: | V-type proton ATPase subunit a, V-type proton ATPase subunit c, V-type proton ATPase subunit c', ... | | 著者 | Mazhab-Jafari, M.T, Rohou, A, Schmidt, C, Bueler, S.A, Benlekbir, S, Robinson, C.V, Rubinstein, J.L. | | 登録日 | 2016-10-03 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase.
Nature, 539, 2016
|
|
3J9U
 
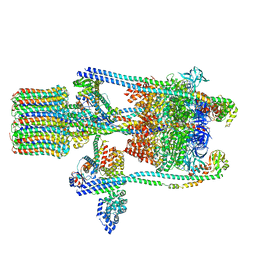 | | Yeast V-ATPase state 2 | | 分子名称: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | 登録日 | 2015-02-23 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7.6 Å) | | 主引用文献 | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3J9T
 
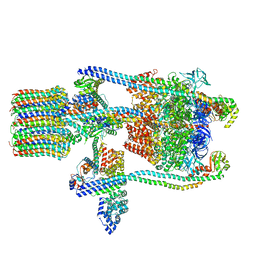 | | Yeast V-ATPase state 1 | | 分子名称: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | 登録日 | 2015-02-23 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
3J9V
 
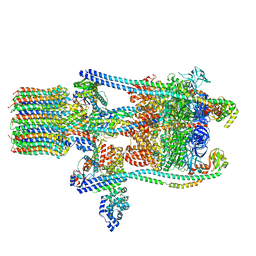 | | Yeast V-ATPase state 3 | | 分子名称: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit C, ... | | 著者 | Zhao, J, Benlekbir, S, Rubinstein, J.L. | | 登録日 | 2015-02-23 | | 公開日 | 2015-05-13 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (8.3 Å) | | 主引用文献 | Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase.
Nature, 521, 2015
|
|
6U7H
 
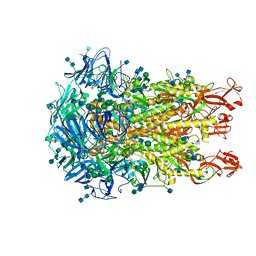 | | Cryo-EM structure of the HCoV-229E spike glycoprotein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Li, Z, Benlekbir, S, Rubinstein, J.L, Rini, J.M. | | 登録日 | 2019-09-02 | | 公開日 | 2019-11-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The human coronavirus HCoV-229E S-protein structure and receptor binding.
Elife, 8, 2019
|
|
8SQL
 
 | |
8SQ0
 
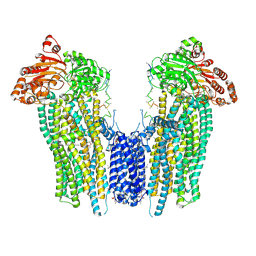 | | Cleaved Ycf1p Dimer in the IFwide-beta conformation | | 分子名称: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, Metal resistance protein YCF1, ... | | 著者 | Bickers, S.C, Benlekbir, S, Rubinstein, J.L, Kanelis, V. | | 登録日 | 2023-05-04 | | 公開日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of a dimeric ABC transporter
To Be Published
|
|
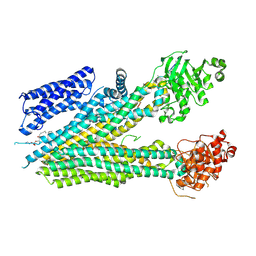
8SQM
 
 | |
7K9Z
 
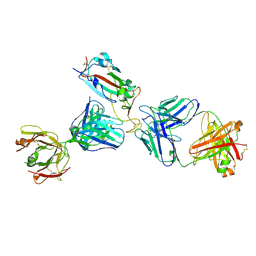 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with the Fab fragments of neutralizing antibodies 298 and 52 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 298 Fab Heavy Chain, 298 Fab Light Chain, ... | | 著者 | Newton, J.C, Kucharska, I, Rujas, E, Cui, H, Julien, J.P. | | 登録日 | 2020-09-29 | | 公開日 | 2020-10-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers.
Nat Commun, 12, 2021
|
|
8U7Z
 
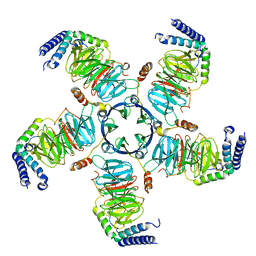 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(CTD)/Gbeta1gamma2 | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U82
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State B From Composite RELION Multi-body Refinement Map | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.84 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U81
 
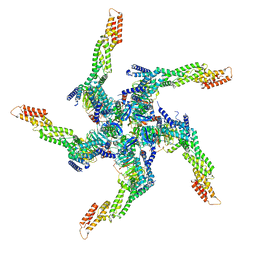 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State A From Composite RELION Multi-body Refinement Map | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.82 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U83
 
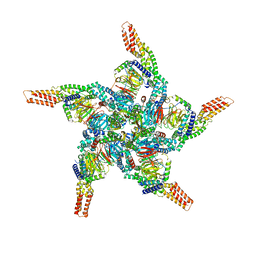 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State C From Composite RELION Multi-body Refinement Map | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.975 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U80
 
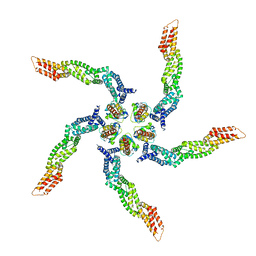 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(BTB)/Cullin3(NTD) | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3 | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U84
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State D From Composite RELION Multi-body Refinement Map | | 分子名称: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | 登録日 | 2023-09-15 | | 公開日 | 2023-10-11 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.88 Å) | | 主引用文献 | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5VKJ
 
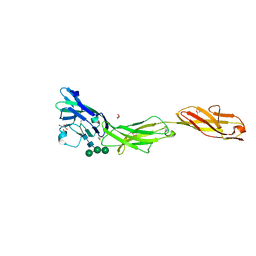 | | Crystal structure of human CD22 Ig domains 1-3 | | 分子名称: | B-cell receptor CD22, GLYCEROL, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Julien, J.P, Ereno-Orbea, J, Sicard, T. | | 登録日 | 2017-04-21 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun, 8, 2017
|
|
5VL3
 
 | | CD22 d1-d3 in complex with therapeutic Fab Epratuzumab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, B-cell receptor CD22, Epratuzumab Fab Heavy Chain, ... | | 著者 | Sicard, T, Ereno-Orbea, J, Julien, J.P. | | 登録日 | 2017-04-24 | | 公開日 | 2017-10-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Molecular basis of human CD22 function and therapeutic targeting.
Nat Commun, 8, 2017
|
|
6OJY
 
 | |
5BXH
 
 | |
