2V6E
 
 | |
1P7D
 
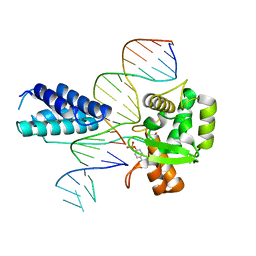 | | Crystal structure of the Lambda Integrase (residues 75-356) bound to DNA | | 分子名称: | 26-MER, 5'-D(*CP*AP*AP*TP*GP*CP*CP*AP*AP*CP*TP*TP*T)-3', Integrase | | 著者 | Aihara, H, Kwon, H.J, Nunes-Duby, S.E, Landy, A, Ellenberger, T. | | 登録日 | 2003-05-01 | | 公開日 | 2003-08-12 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | A Conformational Switch Controls the DNA Cleavage Activity of Lambda Integrase
Mol.Cell, 12, 2003
|
|
1AA3
 
 | | C-TERMINAL DOMAIN OF THE E. COLI RECA, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | RECA | | 著者 | Aihara, H, Ito, Y, Kurumizaka, H, Terada, T, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1997-01-22 | | 公開日 | 1997-07-23 | | 最終更新日 | 2024-04-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing.
J.Mol.Biol., 274, 1997
|
|
1B22
 
 | | RAD51 (N-TERMINAL DOMAIN) | | 分子名称: | DNA REPAIR PROTEIN RAD51 | | 著者 | Aihara, H, Ito, Y, Kurumizaka, H, Yokoyama, S, Shibata, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1998-12-04 | | 公開日 | 1999-12-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The N-terminal domain of the human Rad51 protein binds DNA: structure and a DNA binding surface as revealed by NMR.
J.Mol.Biol., 290, 1999
|
|
6DFY
 
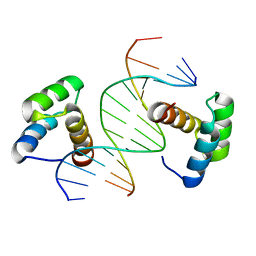 | | Remodeled crystal structure of DNA-bound DUX4-HD2 | | 分子名称: | DNA (5'-D(*AP*AP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*CP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*A)-3'), Double homeobox protein 4 | | 著者 | Aihara, H, Shi, K. | | 登録日 | 2018-05-15 | | 公開日 | 2018-09-05 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.623 Å) | | 主引用文献 | Comment on structural basis of DUX4/IGH-driven transactivation.
Leukemia, 32, 2018
|
|
7R9C
 
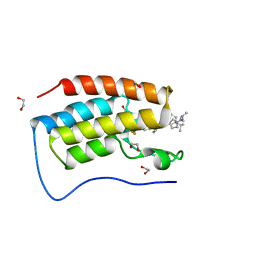 | | Cocrystal of BRD4(D1) with N,N-dimethyl-2-[(3R)-3-(5-{2-[2-methyl-5-(propan-2-yl)phenoxy]pyrimidin-4-yl}-4-[4-(trifluoromethyl)phenyl]-1H-imidazol-1-yl)pyrrolidin-1-yl]ethan-1-amine | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, CHLORIDE ION, ... | | 著者 | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-06-29 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | A Structure-based Design Approach for Generating High Affinity BRD4 D1-Selective Chemical Probes.
J.Med.Chem., 65, 2022
|
|
6E8C
 
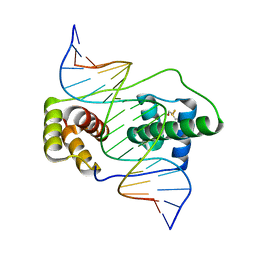 | | Crystal structure of the double homeodomain of DUX4 in complex with DNA | | 分子名称: | DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), Double homeobox protein 4 | | 著者 | Lee, J.K, Bosnakovski, D, Toso, E.A, Dinh, T, Banerjee, S, Bohl, T.E, Shi, K, Kurahashi, K, Kyba, M, Aihara, H. | | 登録日 | 2018-07-27 | | 公開日 | 2018-12-26 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Crystal Structure of the Double Homeodomain of DUX4 in Complex with DNA.
Cell Rep, 25, 2018
|
|
6U82
 
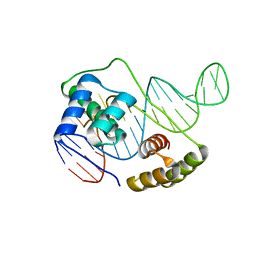 | |
7MC5
 
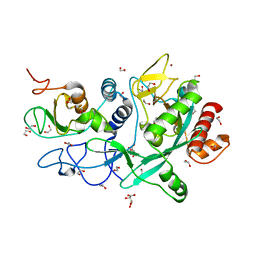 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | 著者 | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | 登録日 | 2021-04-01 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MC6
 
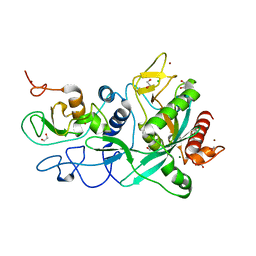 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex containing Mg2+ ion | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | 登録日 | 2021-04-01 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4E10
 
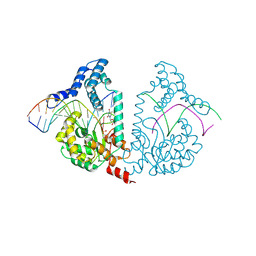 | | Protelomerase tela Y201A covalently complexed with substrate DNA | | 分子名称: | DNA (5'-D(*CP*AP*TP*AP*AP*(BRU)P*AP*AP*CP*AP*AP*(BRU)P*AP*T)-3'), DNA (5'-D(*CP*AP*TP*GP*AP*TP*AP*(BRU)P*(BRU)P*GP*(BRU)P*(BRU)P*AP*(BRU)P*(BRU)P*AP*(BRU)P*G)-3'), Protelomerase, ... | | 著者 | Shi, K, Aihara, H. | | 登録日 | 2012-03-05 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.506 Å) | | 主引用文献 | An enzyme-catalyzed multistep DNA refolding mechanism in hairpin telomere formation.
Plos Biol., 11, 2013
|
|
4NQ3
 
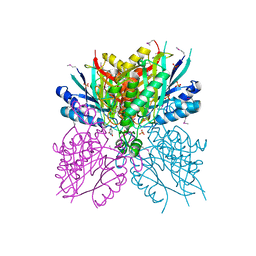 | | Crystal structure of cyanuic acid hydrolase from A. caulinodans | | 分子名称: | BARBITURIC ACID, Cyanuric acid amidohydrolase, MAGNESIUM ION, ... | | 著者 | Cho, S, Shi, K, Aihara, H. | | 登録日 | 2013-11-23 | | 公開日 | 2014-09-10 | | 実験手法 | X-RAY DIFFRACTION (2.702 Å) | | 主引用文献 | Cyanuric acid hydrolase from Azorhizobium caulinodans ORS 571: crystal structure and insights into a new class of Ser-Lys dyad proteins.
Plos One, 9, 2014
|
|
6B0B
 
 | | Crystal structure of human APOBEC3H | | 分子名称: | APOBEC3H, MCherry, RNA (5'-R(*UP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | 著者 | Shaban, N.M, Shi, K, Banerjee, S, Harris, R.S, Aihara, H. | | 登録日 | 2017-09-14 | | 公開日 | 2017-10-25 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.2800622 Å) | | 主引用文献 | The Antiviral and Cancer Genomic DNA Deaminase APOBEC3H Is Regulated by an RNA-Mediated Dimerization Mechanism.
Mol. Cell, 69, 2018
|
|
5W8N
 
 | |
5W8X
 
 | | Lipid A Disaccharide Synthase (LpxB)-7 solubilizing mutations-Bound to UDP | | 分子名称: | Lipid-A-disaccharide synthase, URIDINE-5'-DIPHOSPHATE | | 著者 | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | 登録日 | 2017-06-22 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Nat Commun, 9, 2018
|
|
5WLY
 
 | | E. coli LpxH- 8 mutations | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | 登録日 | 2017-07-28 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The substrate-binding cap of the UDP-diacylglucosamine pyrophosphatase LpxH is highly flexible, enabling facile substrate binding and product release.
J. Biol. Chem., 293, 2018
|
|
5W8S
 
 | | Lipid A Disaccharide Synthase (LpxB)-7 solubilizing mutations | | 分子名称: | LITHIUM ION, Lipid-A-disaccharide synthase, SODIUM ION | | 著者 | Bohl, T.E, Aihara, H, Shi, K, Lee, J.K. | | 登録日 | 2017-06-22 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli.
Nat Commun, 9, 2018
|
|
7TV2
 
 | |
8SBM
 
 | | Crystal structure of the wild-type Catalytic ATP-binding domain of Mtb DosS | | 分子名称: | 1,2-ETHANEDIOL, GAF domain-containing protein, SODIUM ION, ... | | 著者 | Larson, G, Shi, K, Aihara, H, Bhagi-Damodaran, A. | | 登録日 | 2023-04-03 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Understanding ATP Binding to DosS Catalytic Domain with a Short ATP-Lid.
Biochemistry, 62, 2023
|
|
8SPH
 
 | | Crystal structure of chimeric omicron RBD (strain XBB.1) complexed with human ACE2 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, W, Shi, K, Aihara, H, Li, F. | | 登録日 | 2023-05-03 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
8SPI
 
 | | Crystal structure of chimeric omicron RBD (strain XBB.1.5) complexed with human ACE2 | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, W, Shi, K, Aihara, H, Li, F. | | 登録日 | 2023-05-03 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
7MLS
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(2,5-dibromophenoxy)-6-[4-methyl-1-(piperidin-4-yl)-1H-1,2,3-triazol-5-yl]pyridine (compound 23) | | 分子名称: | 1,2-ETHANEDIOL, 2-(2,5-dibromophenoxy)-6-[4-methyl-1-(piperidin-4-yl)-1H-1,2,3-triazol-5-yl]pyridine, Bromodomain-containing protein 4, ... | | 著者 | Cui, H, Johnson, J.A, Zahid, H, Buchholz, C.R, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-04-28 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
7MLR
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(4-{5-[6-(3,5-dimethylphenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1- yl}piperidin-1-yl)-N,N-dimethylethan-1-amine (DW34) | | 分子名称: | 1,2-ETHANEDIOL, 2-(4-{5-[6-(3,5-dimethylphenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine, Bromodomain-containing protein 4, ... | | 著者 | Cui, H, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-04-28 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
7MLQ
 
 | | X-ray crystal structure of human BRD4(D1) in complex with 2-(4-{5-[6-(2,5-dibromophenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine (compound 26) | | 分子名称: | 1,2-ETHANEDIOL, 2-(4-{5-[6-(2,5-dibromophenoxy)pyridin-2-yl]-4-methyl-1H-1,2,3-triazol-1-yl}piperidin-1-yl)-N,N-dimethylethan-1-amine, Bromodomain-containing protein 4, ... | | 著者 | Cui, H, Johnson, J.A, Vail, N.R, Shi, K, Aihara, H, Pomerantz, W.C.K. | | 登録日 | 2021-04-28 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | 4-Methyl-1,2,3-Triazoles as N -Acetyl-Lysine Mimics Afford Potent BET Bromodomain Inhibitors with Improved Selectivity.
J.Med.Chem., 64, 2021
|
|
4YJ0
 
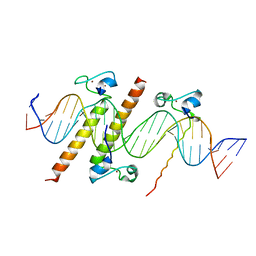 | | Crystal structure of the DM domain of human DMRT1 bound to 25mer target DNA | | 分子名称: | DNA (25-MER), Doublesex- and mab-3-related transcription factor 1, ZINC ION | | 著者 | Murphy, M.W, Lee, J.K, Rojo, S, Gearhart, M.D, Kurahashi, K, Banerjee, S, Loeuille, G, Bashamboo, A, McElreavey, K, Zarkower, D, Aihara, H, Bardwell, V.J. | | 登録日 | 2015-03-02 | | 公開日 | 2015-05-27 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (3.814 Å) | | 主引用文献 | An ancient protein-DNA interaction underlying metazoan sex determination.
Nat.Struct.Mol.Biol., 22, 2015
|
|
