5Y0D
 
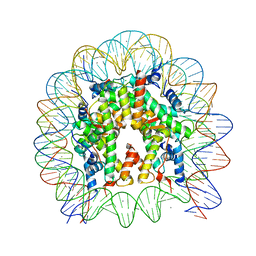 | | Crystal Structure of the human nucleosome containing the H2B E76K mutant | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | 登録日 | 2017-07-16 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
5Y0C
 
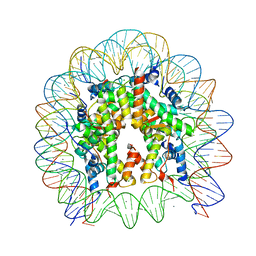 | | Crystal Structure of the human nucleosome at 2.09 angstrom resolution | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Kurumizaka, H, Arimura, Y, Fujita, R, Noda, M. | | 登録日 | 2017-07-16 | | 公開日 | 2018-07-18 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.087 Å) | | 主引用文献 | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
7YOZ
 
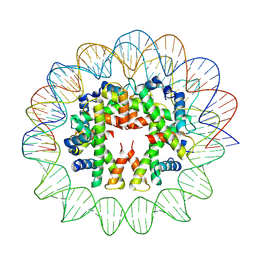 | | Cryo-EM structure of human subnucleosome (intermediate form) | | 分子名称: | Histone H3.1, Histone H4, Widom601 DNA FW (145-MER), ... | | 著者 | Nozawa, K, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2022-08-02 | | 公開日 | 2022-11-16 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cryo-electron microscopy structure of the H3-H4 octasome: A nucleosome-like particle without histones H2A and H2B.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6V2K
 
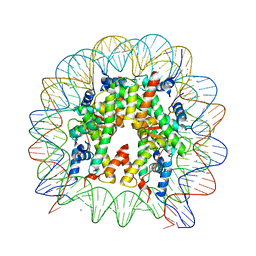 | | The nucleosome structure after H2A-H2B exchange | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A, ... | | 著者 | Arimura, Y, Hirano, R, Kurumizaka, H. | | 登録日 | 2019-11-24 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Histone variant H2A.B-H2B dimers are spontaneously exchanged with canonical H2A-H2B in the nucleosome.
Commun Biol, 4, 2021
|
|
1V5W
 
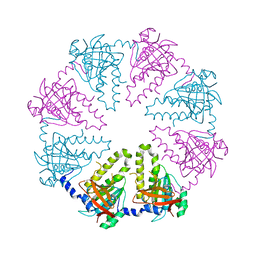 | | Crystal structure of the human Dmc1 protein | | 分子名称: | Meiotic recombination protein DMC1/LIM15 homolog | | 著者 | Kinebuchi, T, Kagawa, W, Enomoto, R, Ikawa, S, Shibata, T, Kurumizaka, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-11-26 | | 公開日 | 2004-05-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein dmc1
Mol.Cell, 14, 2004
|
|
3AZN
 
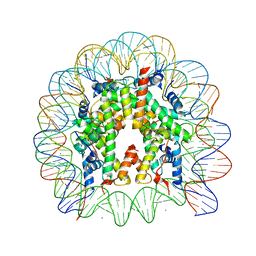 | | Crystal Structure of Human Nucleosome Core Particle Containing H4K91Q mutation | | 分子名称: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | 著者 | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | 登録日 | 2011-05-25 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3AZF
 
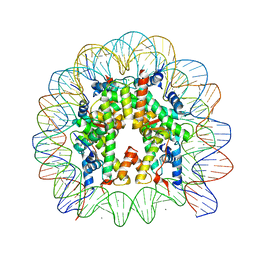 | | Crystal Structure of Human Nucleosome Core Particle Containing H3K79Q mutation | | 分子名称: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | 著者 | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | 登録日 | 2011-05-25 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3AZH
 
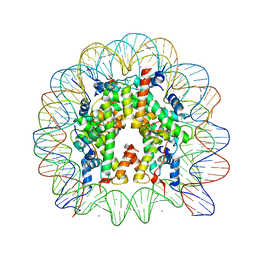 | | Crystal Structure of Human Nucleosome Core Particle Containing H3K122Q mutation | | 分子名称: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | 著者 | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | 登録日 | 2011-05-25 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.49 Å) | | 主引用文献 | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
5XM0
 
 | | The mouse nucleosome structure containing H2A, H2B type3-A, H3.3, and H4 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | 著者 | Taguchi, H, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2017-05-12 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.874 Å) | | 主引用文献 | Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration.
Nat Commun, 9, 2018
|
|
3AZJ
 
 | | Crystal Structure of Human Nucleosome Core Particle Containing H4K44Q mutation | | 分子名称: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | 著者 | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | 登録日 | 2011-05-25 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
3AZK
 
 | | Crystal Structure of Human Nucleosome Core Particle Containing H4K59Q mutation | | 分子名称: | 146-MER DNA, CHLORIDE ION, Histone H2A type 1-B/E, ... | | 著者 | Iwasaki, W, Tachiwana, H, Kawaguchi, K, Shibata, T, Kagawa, W, Kurumizaka, H. | | 登録日 | 2011-05-25 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Comprehensive Structural Analysis of Mutant Nucleosomes Containing Lysine to Glutamine (KQ) Substitutions in the H3 and H4 Histone-Fold Domains
Biochemistry, 50, 2011
|
|
5X7X
 
 | | The crystal structure of the nucleosome containing H3.3 at 2.18 angstrom resolution | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Arimura, Y, Taguchi, H, Kurumizaka, H. | | 登録日 | 2017-02-27 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.184 Å) | | 主引用文献 | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
2EO0
 
 | |
4WD3
 
 | | Crystal structure of an L-amino acid ligase RizA | | 分子名称: | L-amino acid ligase | | 著者 | Kagawa, W, Arai, T, Kino, K, Kurumizaka, H. | | 登録日 | 2014-09-06 | | 公開日 | 2015-09-09 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of RizA, an L-amino-acid ligase from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5XRZ
 
 | | Structure of a ssDNA bound to the inner DNA binding site of RAD52 | | 分子名称: | DNA repair protein RAD52 homolog, POTASSIUM ION, ssDNA (40-MER) | | 著者 | Saotome, M, Saito, K, Yasuda, T, Sugiyama, S, Kurumizaka, H, Kagawa, W. | | 登録日 | 2017-06-11 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural Basis of Homology-Directed DNA Repair Mediated by RAD52
iScience, 3, 2018
|
|
8H0V
 
 | | RNA polymerase II transcribing a chromatosome (type I) | | 分子名称: | DNA (261-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Hirano, R, Ehara, H, Tomoya, K, Takizawa, Y, Sekine, S, Kurumizaka, H. | | 登録日 | 2022-09-30 | | 公開日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of RNA polymerase II transcription on the chromatosome containing linker histone H1.
Nat Commun, 13, 2022
|
|
8H0W
 
 | | RNA polymerase II transcribing a chromatosome (type II) | | 分子名称: | DNA (261-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | 著者 | Hirano, R, Ehara, H, Tomoya, K, Takizawa, Y, Sekine, S, Kurumizaka, H. | | 登録日 | 2022-09-30 | | 公開日 | 2022-12-07 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural basis of RNA polymerase II transcription on the chromatosome containing linker histone H1.
Nat Commun, 13, 2022
|
|
8HE5
 
 | | RNA polymerase II elongation complex bound with Rad26 and Elf1, stalled at SHL(-3.5) of the nucleosome | | 分子名称: | DNA (198-MER), DNA repair protein, DNA-directed RNA polymerase subunit, ... | | 著者 | Osumi, K, Kujirai, T, Ehara, H, Kinoshita, C, Saotome, M, Kagawa, W, Sekine, S, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2022-11-07 | | 公開日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (6.95 Å) | | 主引用文献 | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
1UFI
 
 | | Crystal structure of the dimerization domain of human CENP-B | | 分子名称: | Major centromere autoantigen B | | 著者 | Tawaramoto, M.S, Kurumizaka, H, Tanaka, Y, Park, S.-Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-05-30 | | 公開日 | 2004-02-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of the human centromere protein B (CENP-B) dimerization domain at 1.65-A resolution
J.Biol.Chem., 278, 2003
|
|
5GXQ
 
 | | The crystal structure of the nucleosome containing H3.6 | | 分子名称: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Taguchi, H, Xie, Y, Horikoshi, N, Kurumizaka, H. | | 登録日 | 2016-09-19 | | 公開日 | 2017-04-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
8XBW
 
 | | The cryo-EM structure of the RAD51 N-terminal lobe domain bound to the histone H4 tail of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*CP*CP*GP*CP*TP*TP*AP*AP*AP*CP*GP*CP*AP*CP*GP*TP*A)-3'), DNA (5'-D(P*TP*AP*CP*GP*TP*GP*CP*GP*TP*TP*TP*AP*AP*GP*CP*GP*GP*T)-3'), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBT
 
 | | The cryo-EM structure of the octameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBY
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the blunt end of the nucleosome | | 分子名称: | DNA (5'-D(P*AP*AP*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*GP*TP*T)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.8 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBV
 
 | | The cryo-EM structure of the RAD51 L1 and L2 loops bound to the linker DNA with the sticky end of the nucleosome | | 分子名称: | DNA (5'-D(P*CP*GP*AP*AP*AP*AP*CP*GP*GP*CP*CP*AP*CP*CP*A)-3'), DNA (5'-D(P*TP*GP*GP*CP*CP*GP*TP*TP*TP*TP*CP*G)-3'), DNA repair protein RAD51 homolog 1 | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (7.61 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
8XBU
 
 | | The cryo-EM structure of the decameric RAD51 ring bound to the nucleosome with the linker DNA binding | | 分子名称: | DNA (153-MER), DNA (156-MER), DNA repair protein RAD51 homolog 1, ... | | 著者 | Shioi, T, Hatazawa, S, Ogasawara, M, Takizawa, Y, Kurumizaka, H. | | 登録日 | 2023-12-07 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.24 Å) | | 主引用文献 | Cryo-EM structures of RAD51 assembled on nucleosomes containing a DSB site.
Nature, 628, 2024
|
|
