7YFM
 
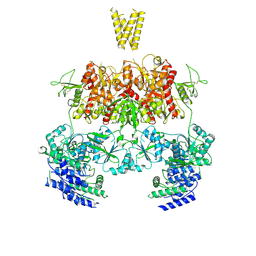 | | Structure of GluN1b-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | 分子名称: | Glutamate receptor ionotropic, NMDA 2D, Isoform 6 of Glutamate receptor ionotropic, ... | | 著者 | Zhang, J.L, Zhu, S.J, Zhang, M. | | 登録日 | 2022-07-08 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFF
 
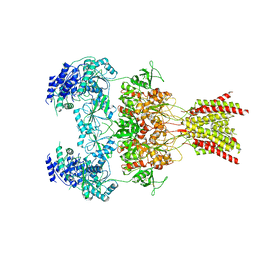 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonist glycine and competitive antagonist CPP. | | 分子名称: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhang, J.L, Zhu, S.J, Zhang, M. | | 登録日 | 2022-07-08 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFL
 
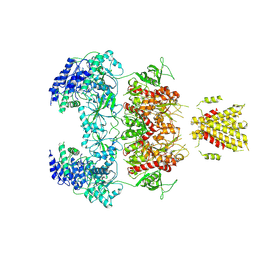 | | Structure of GluN1a-GluN2D NMDA receptor in complex with agonists glycine and glutamate. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | 著者 | Zhang, J.L, Zhu, S.J, Zhang, M. | | 登録日 | 2022-07-08 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7YFO
 
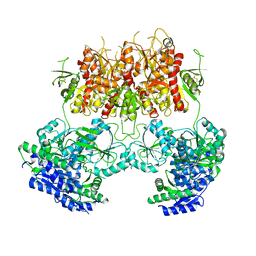 | |
7YFR
 
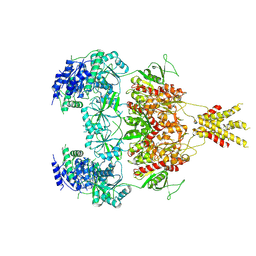 | | Structure of GluN1a E698C-GluN2D NMDA receptor in cystines non-crosslinked state. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | 著者 | Zhang, J.L, Zhu, S.J, Zhang, M. | | 登録日 | 2022-07-09 | | 公開日 | 2023-04-12 | | 最終更新日 | 2023-08-02 | | 実験手法 | ELECTRON MICROSCOPY (5.1 Å) | | 主引用文献 | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2Z60
 
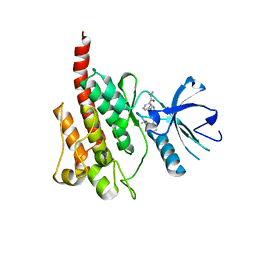 | | Crystal Structure of the T315I Mutant of Abl kinase bound with PPY-A | | 分子名称: | 5-[3-(2-METHOXYPHENYL)-1H-PYRROLO[2,3-B]PYRIDIN-5-YL]-N,N-DIMETHYLPYRIDINE-3-CARBOXAMIDE, Proto-oncogene tyrosine-protein kinase ABL1 | | 著者 | Zhou, T, Dalgarno, D, Zhu, X. | | 登録日 | 2007-07-21 | | 公開日 | 2007-09-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of the T315I Mutant of Abl Kinase
Chem.Biol.Drug Des., 70, 2007
|
|
3R0W
 
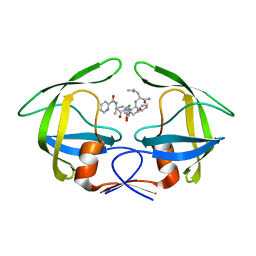 | | Crystal Structures of Multidrug-resistant HIV-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors. | | 分子名称: | Multidrug-resistant clinical isolate 769 HIV-1 Protease, N-[(2R)-1-{[(2S,3S)-5-{[(2R)-1-{[(2S)-1-amino-4-methyl-1-oxopentan-2-yl]amino}-3-chloro-1-oxopropan-2-yl]amino}-3-hydroxy-5-oxo-1-phenylpentan-2-yl]amino}-3-methyl-1-oxobutan-2-yl]pyridine-2-carboxamide | | 著者 | Yedidi, R.S, Gupta, D, Liu, Z, Brunzelle, J, Kovari, I.A, Woster, P.M, Kovari, L.C. | | 登録日 | 2011-03-09 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structures of multidrug-resistant HIV-1 protease in complex with two potent anti-malarial compounds.
Biochem.Biophys.Res.Commun., 421, 2012
|
|
2ZEC
 
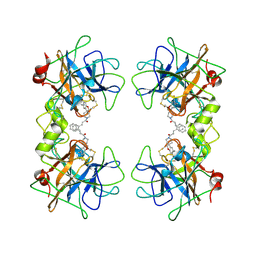 | | Potent, Nonpeptide Inhibitors of Human Mast Cell Tryptase | | 分子名称: | 1-[1'-(3-phenylacryloyl)spiro[1-benzofuran-3,4'-piperidin]-5-yl]methanamine, Tryptase beta 2 | | 著者 | Spurlino, J.C, Lewandowski, F, Milligan, C. | | 登録日 | 2007-12-08 | | 公開日 | 2008-12-09 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.059 Å) | | 主引用文献 | Potent, nonpeptide inhibitors of human mast cell tryptase. Synthesis and biological evaluation of novel spirocyclic piperidine amide derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6INU
 
 | |
2ZIT
 
 | | Structure of the eEF2-ExoA-NAD+ complex | | 分子名称: | Elongation factor 2, Exotoxin A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Jorgensen, R, Merrill, A.R. | | 登録日 | 2008-02-24 | | 公開日 | 2008-06-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The nature and character of the transition state for the ADP-ribosyltransferase reaction.
Embo Rep., 9, 2008
|
|
2ZA5
 
 | | Crystal Structure of human tryptase with potent non-peptide inhibitor | | 分子名称: | (5-(aminomethyl)-2H-spiro[benzofuran-3,4'-piperidine]-1'-yl)(5-(phenylethynyl)furan-2-yl)methanone, Tryptase beta 2 | | 著者 | Spurlino, J.C, Barnakov, S.A, Lewandowski, F, Milligan, C. | | 登録日 | 2007-10-02 | | 公開日 | 2008-02-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Potent, nonpeptide inhibitors of human mast cell tryptase. Synthesis and biological evaluation of novel spirocyclic piperidine amide derivatives
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6INN
 
 | |
6INV
 
 | |
3QUP
 
 | |
6INO
 
 | |
6J30
 
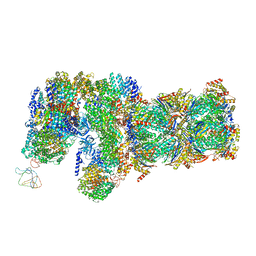 | | yeast proteasome in Ub-engaged state (C2) | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Cong, Y. | | 登録日 | 2019-01-03 | | 公開日 | 2019-03-20 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
6J2N
 
 | | yeast proteasome in substrate-processing state (C3-b) | | 分子名称: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | 著者 | Cong, Y. | | 登録日 | 2019-01-02 | | 公開日 | 2019-03-20 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (7.5 Å) | | 主引用文献 | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
7WK5
 
 | | Cryo-EM structure of Omicron S-ACE2, C2 state | | 分子名称: | Angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Han, W.Y, Wang, Y.F. | | 登録日 | 2022-01-08 | | 公開日 | 2022-02-02 | | 最終更新日 | 2022-03-09 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Molecular basis of SARS-CoV-2 Omicron variant receptor engagement and antibody evasion and neutralization
Biorxiv, 2022
|
|
6J2C
 
 | | Yeast proteasome in translocation competent state (C3-a) | | 分子名称: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | 著者 | Cong, Y. | | 登録日 | 2019-01-01 | | 公開日 | 2019-03-13 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (7 Å) | | 主引用文献 | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
7WK4
 
 | |
7WK6
 
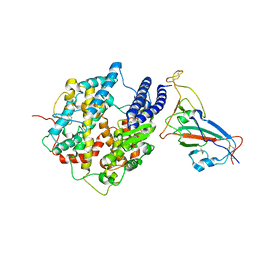 | |
6M39
 
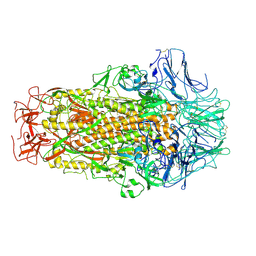 | | Cryo-EM structure of SADS-CoV spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Ouyang, S, Hongxin, G. | | 登録日 | 2020-03-03 | | 公開日 | 2020-08-26 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.55 Å) | | 主引用文献 | Cryo-electron Microscopy Structure of the Swine Acute Diarrhea Syndrome Coronavirus Spike Glycoprotein Provides Insights into Evolution of Unique Coronavirus Spike Proteins.
J.Virol., 94, 2020
|
|
6J2X
 
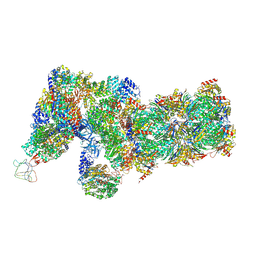 | | Yeast proteasome in resting state (C1-a) | | 分子名称: | 26S PROTEASE REGULATORY SUBUNIT 4 HOMOLOG, 26S PROTEASOME REGULATORY SUBUNIT RPN5, 26S proteasome complex subunit SEM1, ... | | 著者 | Cong, Y. | | 登録日 | 2019-01-03 | | 公開日 | 2019-03-13 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations.
Mol. Cell, 73, 2019
|
|
6IOE
 
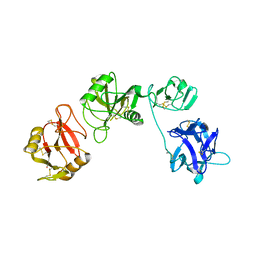 | |
3B8H
 
 | |
