4IP9
 
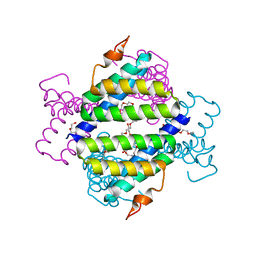 | | Structure of native human serum amyloid A1 | | 分子名称: | O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, Serum amyloid A-1 protein | | 著者 | Lu, J, Sun, P.D. | | 登録日 | 2013-01-09 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural mechanism of serum amyloid A-mediated inflammatory amyloidosis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6VM2
 
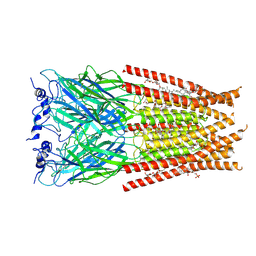 | | Full length Glycine receptor reconstituted in lipid nanodisc in Gly/IVM-conformation (State-2) | | 分子名称: | (2aE,4E,5'S,6S,6'R,7S,8E,11R,13R,15S,17aR,20R,20aR,20bS)-6'-[(2S)-butan-2-yl]-20,20b-dihydroxy-5',6,8,19-tetramethyl-17 -oxo-3',4',5',6,6',10,11,14,15,17,17a,20,20a,20b-tetradecahydro-2H,7H-spiro[11,15-methanofuro[4,3,2-pq][2,6]benzodioxacy clooctadecine-13,2'-pyran]-7-yl 2,6-dideoxy-4-O-(2,6-dideoxy-3-O-methyl-alpha-L-arabino-hexopyranosyl)-3-O-methyl-alpha-L-arabino-hexopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | 著者 | Kumar, A, Basak, S, Chakrapani, S. | | 登録日 | 2020-01-27 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Mechanisms of activation and desensitization of full-length glycine receptor in lipid nanodiscs.
Nat Commun, 11, 2020
|
|
4IP8
 
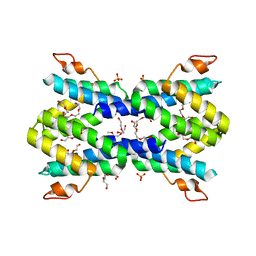 | | Structure of human serum amyloid A1 | | 分子名称: | O-ACETALDEHYDYL-HEXAETHYLENE GLYCOL, SULFATE ION, Serum amyloid A-1 protein | | 著者 | Lu, J, Sun, P.D. | | 登録日 | 2013-01-09 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.193 Å) | | 主引用文献 | Structural mechanism of serum amyloid A-mediated inflammatory amyloidosis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5D8Q
 
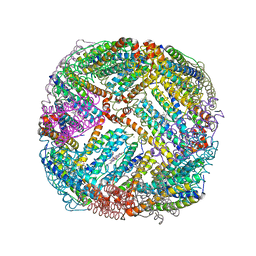 | | 2.20A resolution structure of BfrB (L68A) from Pseudomonas aeruginosa | | 分子名称: | ARSENIC, Ferroxidase, MAGNESIUM ION, ... | | 著者 | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | 登録日 | 2015-08-17 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
5D8O
 
 | | 1.90A resolution structure of BfrB (wild-type, C2221 form) from Pseudomonas aeruginosa | | 分子名称: | Ferroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | 登録日 | 2015-08-17 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
5D8Y
 
 | | 2.05A resolution structure of iron bound BfrB (L68A E81A) from Pseudomonas aeruginosa | | 分子名称: | ACETATE ION, FE (II) ION, Ferroxidase, ... | | 著者 | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | 登録日 | 2015-08-18 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
4OYD
 
 | | Crystal structure of a computationally designed inhibitor of an Epstein-Barr viral Bcl-2 protein | | 分子名称: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, Computationally designed Inhibitor | | 著者 | Shen, B, Procko, E, Baker, D, Stoddard, B. | | 登録日 | 2014-02-11 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A computationally designed inhibitor of an epstein-barr viral bcl-2 protein induces apoptosis in infected cells.
Cell, 157, 2014
|
|
3BWN
 
 | | L-tryptophan aminotransferase | | 分子名称: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, L-tryptophan aminotransferase, PHENYLALANINE, ... | | 著者 | Ferrer, J.-L, Noel, J.P, Pojer, F, Bowman, M, Chory, J, Tao, Y. | | 登録日 | 2008-01-10 | | 公開日 | 2008-04-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants
Cell(Cambridge,Mass.), 133, 2008
|
|
3L57
 
 | |
8B56
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GD-9 | | 分子名称: | (2~{S})-4-(2-chloranylethanoyl)-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5, BROMIDE ION, ... | | 著者 | Straeter, N, Muller, C.E, Claff, T, Sylvester, K, Weisse, R, Gao, S, Song, L, Liu, X, Zhan, P. | | 登録日 | 2022-09-21 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.823 Å) | | 主引用文献 | Discovery and Crystallographic Studies of Nonpeptidic Piperazine Derivatives as Covalent SARS-CoV-2 Main Protease Inhibitors.
J.Med.Chem., 65, 2022
|
|
3EFK
 
 | | Structure of c-Met with pyrimidone inhibitor 50 | | 分子名称: | 5-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-2-[(4-fluorophenyl)amino]-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | 著者 | Bellon, S.F, D'Angelo, N, Whittington, D, Dussault, I. | | 登録日 | 2008-09-09 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Design, synthesis, and biological evaluation of potent c-Met inhibitors.
J.Med.Chem., 51, 2008
|
|
1OGU
 
 | | STRUCTURE OF HUMAN THR160-PHOSPHO CDK2/CYCLIN A COMPLEXED WITH A 2-ARYLAMINO-4-CYCLOHEXYLMETHYL-5-NITROSO-6-AMINOPYRIMIDINE INHIBITOR | | 分子名称: | 4-{[4-AMINO-6-(CYCLOHEXYLMETHOXY)-5-NITROSOPYRIMIDIN-2-YL]AMINO}BENZAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | 著者 | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | 登録日 | 2003-05-13 | | 公開日 | 2003-09-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure-Based Design of 2-Arylamino-4-Cyclohexyl Methyl-5-Nitroso-6-Aminopyrimidine Inhibitors of Cyclin-Dependent Kinases 1 and 2
Bioorg.Med.Chem.Lett., 13, 2003
|
|
6CDX
 
 | | High-resolution crystal structure of fluoropropylated cystine knot, binding to alpha-5 beta-6 integrin | | 分子名称: | cystine knot (fluoropropylated) | | 著者 | Kimura, R, Nix, J, Bongura, C, Chakraborti, S, Gambhir, S, Filipp, F.V. | | 登録日 | 2018-02-09 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Evaluation of integrin alpha v beta6cystine knot PET tracers to detect cancer and idiopathic pulmonary fibrosis.
Nat Commun, 10, 2019
|
|
3EFJ
 
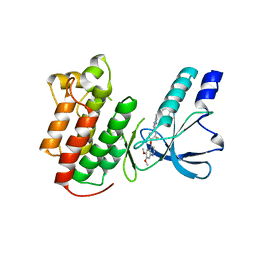 | | Structure of c-Met with pyrimidone inhibitor 7 | | 分子名称: | 2-benzyl-5-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-3-methylpyrimidin-4(3H)-one, Hepatocyte growth factor receptor | | 著者 | D'Angelo, N, Bellon, S, Whittington, D. | | 登録日 | 2008-09-09 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Design, synthesis, and biological evaluation of potent c-Met inhibitors.
J.Med.Chem., 51, 2008
|
|
7CPU
 
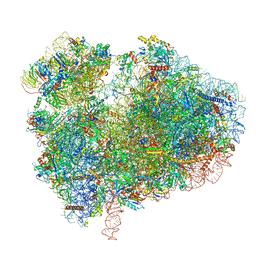 | | Cryo-EM structure of 80S ribosome from mouse kidney | | 分子名称: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | 著者 | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | 登録日 | 2020-08-08 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.82 Å) | | 主引用文献 | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
7CPV
 
 | | Cryo-EM structure of 80S ribosome from mouse testis | | 分子名称: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | 著者 | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | 登録日 | 2020-08-08 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
7CNU
 
 | |
8ACL
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GC-14 | | 分子名称: | (2~{S})-1-(3,4-dichlorophenyl)-4-pyridin-3-ylcarbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Strater, N, Muller, C, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | 登録日 | 2022-07-05 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
8ACD
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GA-17S | | 分子名称: | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Strater, N, Muller, C.E, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | 登録日 | 2022-07-05 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
8IL0
 
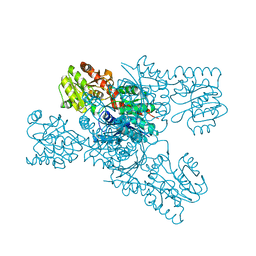 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 | | 分子名称: | Glycosyltransferase | | 著者 | Dai, Y, Li, P, Qiao, H, Xia, M, Liu, W, Fang, P. | | 登録日 | 2023-03-01 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8ILA
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 in complex with substrates | | 分子名称: | (2~{S})-3-[2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-[(1~{R},2~{R})-1-azanyl-2-oxidanyl-propyl]-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-1~{H}-imidazol-5-yl]-2-(trimethyl-$l^{4}-azanyl)propanoic acid, GUANOSINE-5'-DIPHOSPHATE, Glycosyltransferase | | 著者 | Dai, Y, Qiao, H, Xia, M, Fang, P, Liu, W. | | 登録日 | 2023-03-03 | | 公開日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8TBQ
 
 | |
6C2I
 
 | |
4JPO
 
 | | 5A resolution structure of Proteasome Assembly Chaperone Hsm3 in complex with a C-terminal fragment of Rpt1 | | 分子名称: | 26S protease regulatory subunit 7 homolog, DNA mismatch repair protein HSM3 | | 著者 | Lovell, S, Battaile, K.P, Singh, R, Roelofs, J. | | 登録日 | 2013-03-19 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (5 Å) | | 主引用文献 | Reconfiguration of the proteasome during chaperone-mediated assembly.
Nature, 497, 2013
|
|
6CX0
 
 | | Structure of AtTPC1 D376A | | 分子名称: | (1S,3R)-1-(3-{[4-(2-fluorophenyl)piperazin-1-yl]methyl}-4-methoxyphenyl)-2,3,4,9-tetrahydro-1H-beta-carboline-3-carboxylic acid, CALCIUM ION, Two pore calcium channel protein 1 | | 著者 | Kintzer, A.F, Stroud, R.M. | | 登録日 | 2018-04-02 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.501 Å) | | 主引用文献 | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
