5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2017-02-14 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
1T34
 
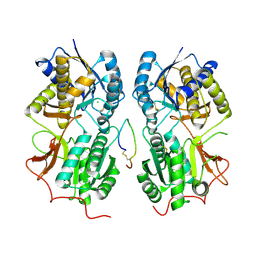 | | ROTATION MECHANISM FOR TRANSMEMBRANE SIGNALING BY THE ATRIAL NATRIURETIC PEPTIDE RECEPTOR | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Atrial natriuretic peptide factor, Atrial natriuretic peptide receptor A, ... | | 著者 | Ogawa, H, Qiu, Y, Ogata, C.M, Misono, K.S. | | 登録日 | 2004-04-23 | | 公開日 | 2004-08-03 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Crystal structure of hormone-bound atrial natriuretic peptide receptor extracellular domain: rotation mechanism for transmembrane signal transduction
J.Biol.Chem., 279, 2004
|
|
4GQ9
 
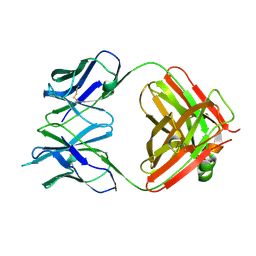 | | Chikungunya virus neutralizing antibody 9.8B Fab fragment | | 分子名称: | Chikungunya virus neutralizing antibody 9.8B Fab fragment heavy chain, Chikungunya virus neutralizing antibody 9.8B Fab fragment light chain | | 著者 | Sun, S, Rossmann, M.G. | | 登録日 | 2012-08-22 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.995 Å) | | 主引用文献 | Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization.
Elife, 2, 2013
|
|
7XQ2
 
 | | Structure of hSLC19A1+2'3'-cGAMP | | 分子名称: | Reduced folate transporter, cGAMP | | 著者 | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | 登録日 | 2022-05-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ1
 
 | | Structure of hSLC19A1+2'3'-CDAS | | 分子名称: | (1~{R},3~{S},6~{R},8~{R},9~{R},10~{S},12~{S},15~{R},17~{R},18~{R})-8,17-bis(6-aminopurin-9-yl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecane-9,18-diol, Reduced folate transporter | | 著者 | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | 登録日 | 2022-05-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XPZ
 
 | | Structure of Apo-hSLC19A1 | | 分子名称: | Reduced folate transporter | | 著者 | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | 登録日 | 2022-05-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
7XQ0
 
 | | Structure of hSLC19A1+3'3'-CDA | | 分子名称: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Reduced folate transporter | | 著者 | Zhang, Q.X, Zhang, X.Y, Zhu, Y.L, Sun, P.P, Gao, A, Zhang, L.G, Gao, P. | | 登録日 | 2022-05-06 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Recognition of cyclic dinucleotides and folates by human SLC19A1.
Nature, 612, 2022
|
|
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | 著者 | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2017-02-14 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
7YLM
 
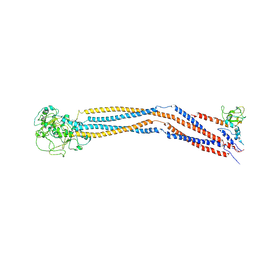 | | Cryo-EM structure of 8-subunit Smc5/6 hinge region | | 分子名称: | MMS21 isoform 1, SMC6 isoform 1, Structural maintenance of chromosomes protein 5 | | 著者 | Qian, L, Jun, Z, Xiang, Z, Wang, Z, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | 登録日 | 2022-07-26 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (6.17 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
4HRE
 
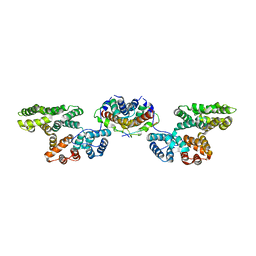 | | Crystal Structure of p11/Annexin A2 Heterotetramer in Complex with SMARCA3 Peptide | | 分子名称: | Annexin A2, Helicase-like transcription factor, Protein S100-A10 | | 著者 | Gao, P, Patel, D.J. | | 登録日 | 2012-10-27 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.7852 Å) | | 主引用文献 | SMARCA3, a Chromatin-Remodeling Factor, Is Required for p11-Dependent Antidepressant Action.
Cell(Cambridge,Mass.), 152, 2013
|
|
6AKK
 
 | | Crystal structure of the second Coiled-coil domain of SIKE1 | | 分子名称: | GLYCEROL, Suppressor of IKBKE 1 | | 著者 | Zhou, L, Chen, M, Zhou, Z.C. | | 登録日 | 2018-09-02 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
4I0S
 
 | |
6AKL
 
 | | Crystal structure of Striatin3 in complex with SIKE1 Coiled-coil domain | | 分子名称: | Striatin-3, Suppressor of IKBKE 1 | | 著者 | Zhou, L, Chen, M, Zhou, Z.C. | | 登録日 | 2018-09-02 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
4NNH
 
 | |
7VOJ
 
 | | Al-bound structure of the AtALMT1 mutant M60A | | 分子名称: | ACETIC ACID, ALUMINUM ION, Aluminum-activated malate transporter 1 | | 著者 | Wang, J. | | 登録日 | 2021-10-14 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of ALMT1-mediated aluminum resistance in Arabidopsis.
Cell Res., 32, 2022
|
|
7VQ5
 
 | |
7VQ3
 
 | |
7VQ4
 
 | |
7VQ7
 
 | |
6IDV
 
 | | Peptide Asparaginyl Ligases from Viola yedoensis | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | El Sahili, A, Hu, S, Lescar, J. | | 登録日 | 2018-09-11 | | 公開日 | 2019-05-15 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural determinants for peptide-bond formation by asparaginyl ligases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7E0F
 
 | |
7EKQ
 
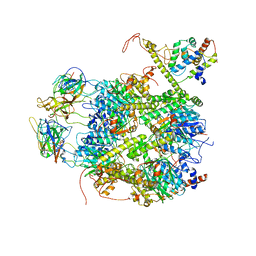 | | CrClpP-S2c | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit, ... | | 著者 | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | 登録日 | 2021-04-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7EKO
 
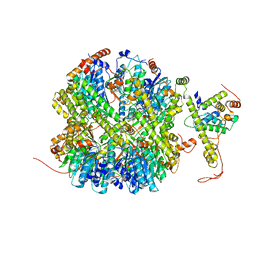 | | CrClpP-S1 | | 分子名称: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit | | 著者 | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | 登録日 | 2021-04-06 | | 公開日 | 2021-10-20 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
4MPZ
 
 | |
7ENS
 
 | |
