6VPR
 
 | |
6VIK
 
 | |
6TF1
 
 | |
6TFF
 
 | |
6TF0
 
 | | Crystal structure of the ADP-binding domain of the NAD+ riboswitch with Nicotinamide adenine dinucleotide, reduced (NADH) | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Chains: A, MAGNESIUM ION, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2019-11-12 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and ligand binding of the ADP-binding domain of the NAD+ riboswitch.
Rna, 26, 2020
|
|
6TF3
 
 | |
6TFE
 
 | |
5YK3
 
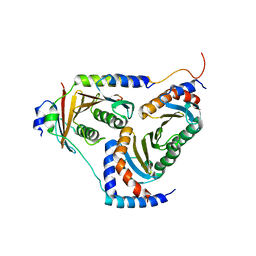 | | human Ragulator complex | | 分子名称: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | 著者 | Wu, G, Mu, Z. | | 登録日 | 2017-10-12 | | 公開日 | 2019-01-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Structural insight into the Ragulator complex which anchors mTORC1 to the lysosomal membrane
Cell Discov, 3, 2017
|
|
6VIH
 
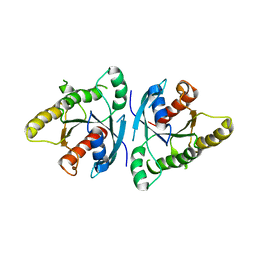 | |
4CKR
 
 | | Crystal structure of the human DDR1 kinase domain in complex with DDR1-IN-1 | | 分子名称: | 1,2-ETHANEDIOL, 4-[(4-ethylpiperazin-1-yl)methyl]-n-{4-methyl-3-[(2-oxo-2,3-dihydro-1h-indol-5-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, EPITHELIAL DISCOIDIN DOMAIN-CONTAINING RECEPTOR 1 | | 著者 | Canning, P, Elkins, J.M, Goubin, S, Mahajan, P, Krojer, T, Newman, J.A, Dixon-Clarke, S, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | 登録日 | 2014-01-07 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Discovery of a Potent and Selective Ddr1 Receptor Tyrosine Kinase Inhibitor.
Acs Chem.Biol., 8, 2013
|
|
7WRO
 
 | |
7WR8
 
 | |
7WRL
 
 | |
7WRZ
 
 | |
5U09
 
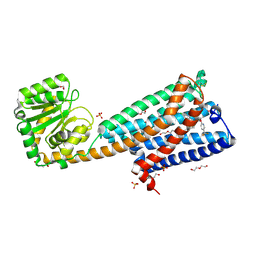 | | High-resolution crystal structure of the human CB1 cannabinoid receptor | | 分子名称: | Cannabinoid receptor 1,GlgA glycogen synthase, DI(HYDROXYETHYL)ETHER, N-[(2S,3S)-4-(4-chlorophenyl)-3-(3-cyanophenyl)butan-2-yl]-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]oxy}propanamide, ... | | 著者 | Shao, Z.H, Yin, J, Rosenbaum, D. | | 登録日 | 2016-11-23 | | 公開日 | 2016-12-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | High-resolution crystal structure of the human CB1 cannabinoid receptor.
Nature, 2016
|
|
6K8K
 
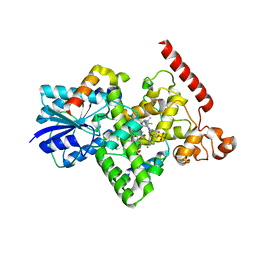 | | Crystal structure of Arabidopsis thaliana BIC2-CRY2 complex | | 分子名称: | ADENOSINE MONOPHOSPHATE, Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Wang, X, Ma, L, Guan, Z, Yin, P. | | 登録日 | 2019-06-12 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4M9X
 
 | | Crystal structure of CED-4 bound CED-3 fragment | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | 著者 | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | 登録日 | 2013-08-15 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.344 Å) | | 主引用文献 | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
1BJ9
 
 | | EFFECT OF UNNATURAL HEME SUBSTITUTION ON KINETICS OF ELECTRON TRANSFER IN CYTOCHROME C PEROXIDASE | | 分子名称: | CYTOCHROME C PEROXIDASE, [7,12-DEACETYL-3,8,13,17-TETRAMETHYL-21H,23H-PORPHINE-2,18-DIPROPANOATO(2-)-N21,N22,N23,N24]-IRON | | 著者 | Miller, M.A, Kraut, J. | | 登録日 | 1998-07-03 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Effect of Unnatural Heme Substitution on Kinetics of Electron Transfer in Cytochrome C Peroxidase
To be Published
|
|
4C2I
 
 | | Cryo-EM structure of Dengue virus serotype 1 complexed with Fab fragments of human antibody 1F4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Fibriansah, G, Tan, J.L, de Alwis, R, Smith, S.A, Ng, T.-S, Kostyuchenko, V.A, Ibarra, K.D, Harris, E, de Silva, A, Crowe Junior, J.E, Lok, S.-M. | | 登録日 | 2013-08-18 | | 公開日 | 2014-01-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | A Potent Anti-Dengue Human Antibody Preferentially Recognizes the Conformation of E Protein Monomers Assembled on the Virus Surface.
Embo Mol.Med., 6, 2014
|
|
4M9Y
 
 | | Crystal structure of CED-4 bound CED-3 fragment | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | 著者 | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | 登録日 | 2013-08-15 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (4.2 Å) | | 主引用文献 | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
6JEA
 
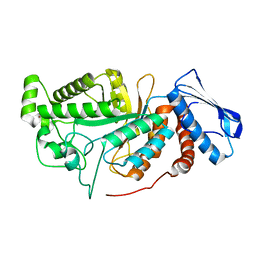 | | crystal structure of a beta-N-acetylhexosaminidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ZINC ION | | 著者 | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | 登録日 | 2019-02-04 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.275 Å) | | 主引用文献 | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
3A2O
 
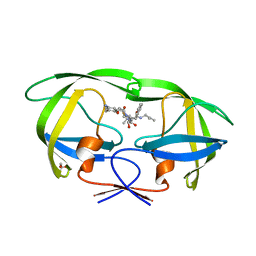 | | Crystal Structure of HIV-1 Protease Complexed with KNI-1689 | | 分子名称: | (4R)-3-[(2S,3S)-3-{[(4-amino-2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-5,5-dimethyl-N-(2-methylprop -2-en-1-yl)-1,3-thiazolidine-4-carboxamide, GLYCEROL, PROTEASE | | 著者 | Adachi, M, Tamada, T, Hidaka, K, Kimura, T, Kiso, Y, Kuroki, R. | | 登録日 | 2009-05-26 | | 公開日 | 2010-03-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (0.88 Å) | | 主引用文献 | Small-sized human immunodeficiency virus type-1 protease inhibitors containing allophenylnorstatine to explore the S2' pocket.
J.Med.Chem., 52, 2009
|
|
6JE8
 
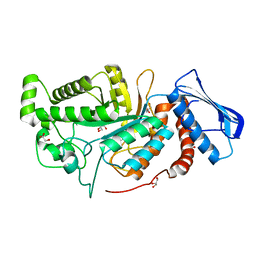 | | crystal structure of a beta-N-acetylhexosaminidase | | 分子名称: | Beta-N-acetylhexosaminidase, FORMIC ACID, GLYCEROL, ... | | 著者 | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | 登録日 | 2019-02-04 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.798 Å) | | 主引用文献 | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
6JIV
 
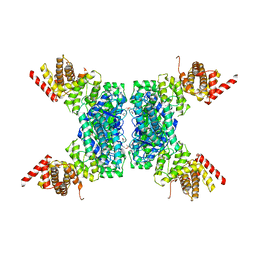 | | SspE crystal structure | | 分子名称: | SspE protein | | 著者 | Bing, Y.Z, Yang, H.G. | | 登録日 | 2019-02-23 | | 公開日 | 2020-03-25 | | 最終更新日 | 2020-07-08 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | SspABCD-SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities.
Nat Microbiol, 5, 2020
|
|
6JEB
 
 | | crystal structure of a beta-N-acetylhexosaminidase | | 分子名称: | ACETAMIDE, Beta-N-acetylhexosaminidase, ZINC ION | | 著者 | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | 登録日 | 2019-02-05 | | 公開日 | 2019-03-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.498 Å) | | 主引用文献 | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
