6E5U
 
 | |
4LGI
 
 | | N-terminal truncated NleC structure | | 分子名称: | Uncharacterized protein | | 著者 | Li, W.Q, Liu, Y.X, Sheng, X.L, Yan, C.Y, Wang, J.W. | | 登録日 | 2013-06-28 | | 公開日 | 2014-01-15 | | 最終更新日 | 2015-02-25 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Structure and mechanism of a type III secretion protease, NleC
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5XNE
 
 | |
6LBH
 
 | |
2K3K
 
 | |
6TFG
 
 | |
4LGJ
 
 | | Crystal structure and mechanism of a type III secretion protease | | 分子名称: | Uncharacterized protein, ZINC ION | | 著者 | Li, W.Q, Liu, Y.X, Sheng, X.L, Yan, C.Y, Wang, J.W. | | 登録日 | 2013-06-28 | | 公開日 | 2014-01-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure and mechanism of a type III secretion protease, NleC
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6ULY
 
 | | Adenylation domain of a keto acid-selecting NRPS module bound to keto acyl adenylate space group P212121 | | 分子名称: | 5'-O-{(S)-hydroxy[(4-methyl-2-oxopentanoyl)oxy]phosphoryl}adenosine, Amino acid adenylation domain-containing protein, GLYCEROL, ... | | 著者 | Alonzo, D.A, Chiche-Lapierre, C, Schmeing, T.M. | | 登録日 | 2019-10-08 | | 公開日 | 2020-02-19 | | 最終更新日 | 2020-05-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis of keto acid utilization in nonribosomal depsipeptide synthesis.
Nat.Chem.Biol., 16, 2020
|
|
6VAC
 
 | | Mouse retromer (VPS26/VPS35/VPS29) heterotrimer | | 分子名称: | Vacuolar protein sorting-associated protein 26A, Vacuolar protein sorting-associated protein 29, Vacuolar protein sorting-associated protein 35 | | 著者 | Kendall, A.K, Jackson, L.P. | | 登録日 | 2019-12-17 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (5.7 Å) | | 主引用文献 | Mammalian Retromer Is an Adaptable Scaffold for Cargo Sorting from Endosomes.
Structure, 28, 2020
|
|
6TFH
 
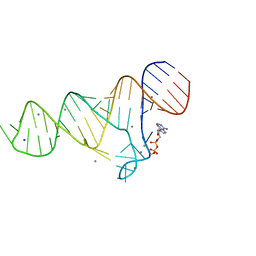 | |
4M9Z
 
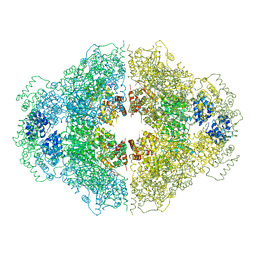 | | Crystal structure of CED-4 bound CED-3 fragment | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | 著者 | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | 登録日 | 2013-08-15 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.405 Å) | | 主引用文献 | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
6US8
 
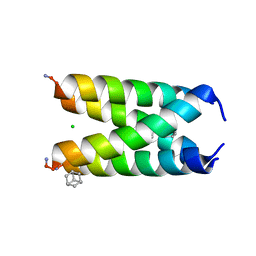 | |
7BP3
 
 | | Cryo-EM structure of the human MCT2 | | 分子名称: | Monocarboxylate transporter 2 | | 著者 | Zhang, B, Jin, Q, Zhang, X, Guo, J, Ye, S. | | 登録日 | 2020-03-21 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cooperative transport mechanism of human monocarboxylate transporter 2.
Nat Commun, 11, 2020
|
|
6M6I
 
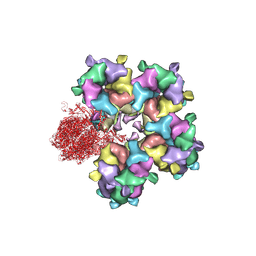 | | Structure of HSV2 B-capsid portal vertex | | 分子名称: | Coiled coils chain 1, Coiled coils chain 2, Major capsid protein, ... | | 著者 | Wang, X.X, Wang, N. | | 登録日 | 2020-03-14 | | 公開日 | 2021-03-10 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6H
 
 | | Structure of HSV2 C-capsid portal vertex | | 分子名称: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | 著者 | Wang, X.X, Wang, N. | | 登録日 | 2020-03-14 | | 公開日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6G
 
 | | Structure of HSV2 viron capsid portal vertex | | 分子名称: | Capsid vertex component 1, Capsid vertex component 2, Coiled coils, ... | | 著者 | Wang, X.X, Wang, N. | | 登録日 | 2020-03-14 | | 公開日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (5.39 Å) | | 主引用文献 | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
4BEW
 
 | | SERCA bound to phosphate analogue | | 分子名称: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, ACETATE ION, MAGNESIUM ION, ... | | 著者 | Drachmann, N.D, Mattle, D, Laursen, M, Bublitz, M, Olesen, C, Moeller, J.V, Nissen, P, Morth, J.P. | | 登録日 | 2013-03-12 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Serca Bound to Phosphate Analogue
To be Published
|
|
6US9
 
 | |
6LNM
 
 | |
6VAB
 
 | |
5XYB
 
 | |
6VII
 
 | |
5Y24
 
 | | Crystal structure of AimR from Bacillus phage SPbeta in complex with its signalling peptide | | 分子名称: | AimR transcriptional regulator, BROMIDE ION, GLY-MET-PRO-ARG-GLY-ALA | | 著者 | Wang, Q, Guan, Z.Y, Zou, T.T, Yin, P. | | 登録日 | 2017-07-24 | | 公開日 | 2018-09-19 | | 最終更新日 | 2018-11-28 | | 実験手法 | X-RAY DIFFRACTION (1.922 Å) | | 主引用文献 | Structural basis of the arbitrium peptide-AimR communication system in the phage lysis-lysogeny decision.
Nat Microbiol, 3, 2018
|
|
5HBW
 
 | | RNA primer-template complex with 2-methylimidazole-activated monomer analogue | | 分子名称: | RNA (5'-R(*(LCC)P*(TLN)P*(LCG)P*UP*AP*CP*A)-3'), [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-(3-methyl-1~{H}-pyrazol-4-yl)phosphinic acid | | 著者 | Zhang, W, Tam, C.P, Szostak, J.W. | | 登録日 | 2016-01-03 | | 公開日 | 2016-12-07 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Unusual Base-Pairing Interactions in Monomer-Template Complexes.
ACS Cent Sci, 2, 2016
|
|
4M9S
 
 | | crystal structure of CED-4 bound CED-3 fragment | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CED-3 fragment, Cell death protein 4, ... | | 著者 | Huang, W.J, Jinag, T.Y, Choi, W.Y, Wang, J.W, Shi, Y.G. | | 登録日 | 2013-08-15 | | 公開日 | 2013-10-23 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (3.207 Å) | | 主引用文献 | Mechanistic insights into CED-4-mediated activation of CED-3.
Genes Dev., 27, 2013
|
|
