3HD4
 
 | | MHV Nucleocapsid Protein NTD | | 分子名称: | Nucleoprotein | | 著者 | Giedroc, D.P, Keane, S.C, Dann III, C.E. | | 登録日 | 2009-05-06 | | 公開日 | 2009-11-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.747 Å) | | 主引用文献 | Coronavirus N Protein N-Terminal Domain (NTD) Specifically Binds the Transcriptional Regulatory Sequence (TRS) and Melts TRS-cTRS RNA Duplexes.
J.Mol.Biol., 394, 2009
|
|
3QFL
 
 | |
8KHQ
 
 | | Bifunctional sulfoxide synthase OvoA_Th2 in complex with histidine and cysteine | | 分子名称: | 5-histidylcysteine sulfoxide synthase/putative 4-mercaptohistidine N1-methyltranferase, COBALT (II) ION, CYSTEINE, ... | | 著者 | Wang, J, Ye, K, Wang, X.Y, Yan, W.P. | | 登録日 | 2023-08-22 | | 公開日 | 2023-12-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.69 Å) | | 主引用文献 | Biochemical and Structural Characterization of OvoA Th2 : A Mononuclear Nonheme Iron Enzyme from Hydrogenimonas thermophila for Ovothiol Biosynthesis.
Acs Catalysis, 13, 2023
|
|
4Y5T
 
 | | Structure of FtmOx1 apo with metal Iron | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COBALT (II) ION, FE (II) ION, ... | | 著者 | Yan, W, Zhang, Y. | | 登録日 | 2015-02-12 | | 公開日 | 2015-11-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Endoperoxide formation by an alpha-ketoglutarate-dependent mononuclear non-haem iron enzyme.
Nature, 527, 2015
|
|
1W24
 
 | | Crystal Structure Of human Vps29 | | 分子名称: | VACUOLAR PROTEIN SORTING PROTEIN 29 | | 著者 | Wang, D, Guo, M, Teng, M, Niu, L. | | 登録日 | 2004-06-26 | | 公開日 | 2005-03-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of Human Vacuolar Protein Sorting Protein 29 Reveals a Phosphodiesterase/Nuclease-Like Fold and Two Protein-Protein Interaction Sites.
J.Biol.Chem., 280, 2005
|
|
4Y5S
 
 | |
4Y86
 
 | | Crystal structure of PDE9 in complex with racemic inhibitor C33 | | 分子名称: | 6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, 6-{[(1S)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, ... | | 著者 | Huang, M. | | 登録日 | 2015-02-16 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
4Y87
 
 | | Crystal structure of phosphodiesterase 9 in complex with (R)-C33 (6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one) | | 分子名称: | 6-{[(1R)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | 著者 | Huang, M. | | 登録日 | 2015-02-16 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
4Y8C
 
 | | Crystal structure of phosphodiesterase 9 in complex with (S)-C33 | | 分子名称: | 6-{[(1S)-1-(4-chlorophenyl)ethyl]amino}-1-cyclopentyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | 著者 | Huang, M. | | 登録日 | 2015-02-16 | | 公開日 | 2015-09-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural Asymmetry of Phosphodiesterase-9A and a Unique Pocket for Selective Binding of a Potent Enantiomeric Inhibitor.
Mol.Pharmacol., 88, 2015
|
|
4HPO
 
 | | Crystal structure of RV144-elicited antibody CH58 in complex with V2 peptide | | 分子名称: | CH58 Fab heavy chain, CH58 Fab light chain, Envelope glycoprotein gp160, ... | | 著者 | McLellan, J.S, Gorman, J, Haynes, B.F, Kwong, P.D. | | 登録日 | 2012-10-24 | | 公開日 | 2013-02-06 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.694 Å) | | 主引用文献 | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
4HQQ
 
 | |
7WSB
 
 | | The ternary complex structure of FtmOx1 with a-ketoglutarate and 13-oxo-fumitremorgin B | | 分子名称: | 13-Oxofumitremorgin B, 2-OXOGLUTARIC ACID, COBALT (II) ION, ... | | 著者 | Wang, J, Wang, X.Y, Wang, Y.Y, Yan, W.P. | | 登録日 | 2022-01-28 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Dissecting the Mechanism of the Nonheme Iron Endoperoxidase FtmOx1 Using Substrate Analogues.
Jacs Au, 2, 2022
|
|
4HPY
 
 | | Crystal structure of RV144-elicited antibody CH59 in complex with V2 peptide | | 分子名称: | CH59 Fab heavy chain, CH59 Fab light chain, Envelope glycoprotein gp160, ... | | 著者 | McLellan, J.S, Gorman, J, Haynes, B.F, Kwong, P.D. | | 登録日 | 2012-10-24 | | 公開日 | 2013-02-06 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Vaccine Induction of Antibodies against a Structurally Heterogeneous Site of Immune Pressure within HIV-1 Envelope Protein Variable Regions 1 and 2.
Immunity, 38, 2013
|
|
6M4O
 
 | | Cryo-EM structure of the monomeric SPT-ORMDL3 complex | | 分子名称: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | 著者 | Li, S.S, Xie, T, Wang, L, Gong, X. | | 登録日 | 2020-03-08 | | 公開日 | 2021-02-10 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6M4N
 
 | | Cryo-EM structure of the dimeric SPT-ORMDL3 complex | | 分子名称: | ORM1-like protein 3, PYRIDOXAL-5'-PHOSPHATE, Serine palmitoyltransferase 1, ... | | 著者 | Li, S.S, Xie, T, Wang, L, Gong, X. | | 登録日 | 2020-03-07 | | 公開日 | 2021-02-10 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural insights into the assembly and substrate selectivity of human SPT-ORMDL3 complex.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6N9T
 
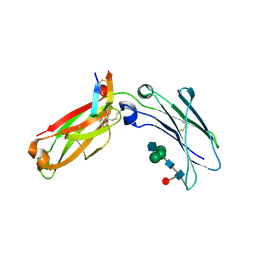 | | Structure of a peptide-based photo-affinity cross-linker with Herceptin Fc | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin G1 FC, ... | | 著者 | Sadowsky, J, Ultsch, M, Vance, N, Wang, W. | | 登録日 | 2018-12-04 | | 公開日 | 2019-01-16 | | 最終更新日 | 2025-04-02 | | 実験手法 | X-RAY DIFFRACTION (2.576 Å) | | 主引用文献 | Development, Optimization, and Structural Characterization of an Efficient Peptide-Based Photoaffinity Cross-Linking Reaction for Generation of Homogeneous Conjugates from Wild-Type Antibodies.
Bioconjug. Chem., 30, 2019
|
|
6O6L
 
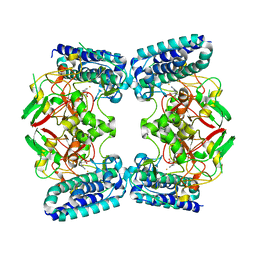 | | The Structure of EgtB(Cabther) in complex with Hercynine | | 分子名称: | EgtB (Cabther), FE (III) ION, N,N,N-trimethyl-histidine | | 著者 | Irani, S, Zhang, Y. | | 登録日 | 2019-03-07 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Crystal Structure of the Ergothioneine Sulfoxide Synthase fromCandidatus Chloracidobacterium thermophilumand Structure-Guided Engineering To Modulate Its Substrate Selectivity.
Acs Catalysis, 9, 2019
|
|
7XDQ
 
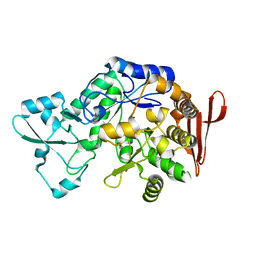 | | Crystal structure of a glucosylglycerol phosphorylase mutant from Marinobacter adhaerens | | 分子名称: | Glucosylglycerol phosphorylase, LITHIUM ION, beta-D-glucopyranose | | 著者 | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | 登録日 | 2022-03-28 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
7XDR
 
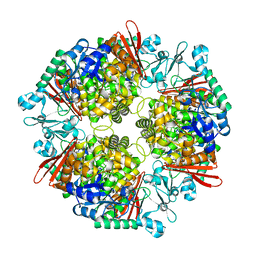 | | Crystal structure of a glucosylglycerol phosphorylase from Marinobacter adhaerens | | 分子名称: | Glucosylglycerol phosphorylase | | 著者 | Wei, H.L, Li, Q, Yang, J.G, Liu, W.D, Sun, Y.X. | | 登録日 | 2022-03-28 | | 公開日 | 2023-02-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Protein Engineering of Glucosylglycerol Phosphorylase Facilitating Efficient and Highly Regio- and Stereoselective Glycosylation of Polyols in a Synthetic System.
Acs Catalysis, 2022
|
|
8ZUF
 
 | | Cryo-EM structure of P.nat ACE2 mutant in complex with MOW15-22 RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Tang, J, Deng, Z. | | 登録日 | 2024-06-09 | | 公開日 | 2025-02-12 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Multiple independent acquisitions of ACE2 usage in MERS-related coronaviruses.
Cell, 188, 2025
|
|
7DZH
 
 | | intermediate of FABP with a delay time of 100 ns | | 分子名称: | Fatty acid-binding protein, liver, PALMITIC ACID | | 著者 | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | 登録日 | 2021-01-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZE
 
 | | Fabp ground state captured by XFELs | | 分子名称: | Fatty acid-binding protein, liver, PALMITIC ACID | | 著者 | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | 登録日 | 2021-01-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZF
 
 | | Intermediate of FABP with a delay time of 10 ns | | 分子名称: | Fatty acid-binding protein, liver, PALMITIC ACID | | 著者 | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | 登録日 | 2021-01-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZG
 
 | | Intermediate of FABP with a delay time of 30 ns | | 分子名称: | Fatty acid-binding protein, liver, PALMITIC ACID | | 著者 | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | 登録日 | 2021-01-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZI
 
 | | intermediate of FABP with a delay time of 300 ns | | 分子名称: | Fatty acid-binding protein, liver, PALMITIC ACID | | 著者 | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | 登録日 | 2021-01-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
