6R5I
 
 | |
6R5P
 
 | |
6R5U
 
 | |
2JTN
 
 | | NMR Solution Structure of a ldb1-LID:Lhx3-LIM complex | | 分子名称: | LIM domain-binding protein 1, LIM/homeobox protein Lhx3, ZINC ION | | 著者 | Lee, C, Nancarrow, A.L, Mackay, J.P, Matthews, J.M. | | 登録日 | 2007-08-03 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes
Embo J., 27, 2008
|
|
7LRQ
 
 | | Crystal structure of human SFPQ/NONO heterodimer, conserved DBHS region | | 分子名称: | CHLORIDE ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | 著者 | Marshall, A.C, Bond, C.S, Mohnen, I. | | 登録日 | 2021-02-17 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Paraspeckle subnuclear bodies depend on dynamic heterodimerisation of DBHS RNA-binding proteins via their structured domains.
J.Biol.Chem., 298, 2022
|
|
6Z1X
 
 | |
6Z1W
 
 | |
5LY7
 
 | | Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | 分子名称: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Beta-hexosaminidase, DI(HYDROXYETHYL)ETHER | | 著者 | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | 登録日 | 2016-09-25 | | 公開日 | 2017-05-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
4PD6
 
 | |
4PD8
 
 | | Structure of vcCNT-7C8C bound to pyrrolo-cytidine | | 分子名称: | 6-methyl-3-(beta-D-ribofuranosyl)-3,7-dihydro-2H-pyrrolo[2,3-d]pyrimidin-2-one, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | 著者 | Johnson, Z.L, Lee, S.-Y. | | 登録日 | 2014-04-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
2IXU
 
 | |
2J8F
 
 | |
2IXV
 
 | |
4PB1
 
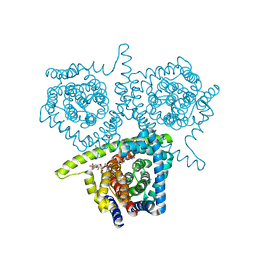 | | Structure of vcCNT-7C8C bound to ribavirin | | 分子名称: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | 著者 | Johnson, Z.L, Lee, S.-Y. | | 登録日 | 2014-04-11 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.803 Å) | | 主引用文献 | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD5
 
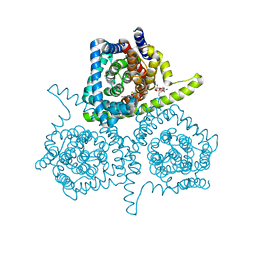 | |
4PDA
 
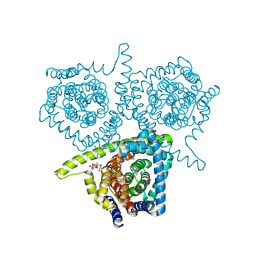 | | Structure of vcCNT-7C8C bound to cytidine | | 分子名称: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | 著者 | Johnson, Z.L, Lee, S.-Y. | | 登録日 | 2014-04-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.608 Å) | | 主引用文献 | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4PD9
 
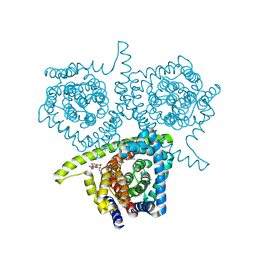 | | Structure of vcCNT-7C8C bound to adenosine | | 分子名称: | ADENOSINE, DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, ... | | 著者 | Johnson, Z.L, Lee, S.-Y. | | 登録日 | 2014-04-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.096 Å) | | 主引用文献 | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
4CJN
 
 | | Crystal structure of PBP2a from MRSA in complex with quinazolinone ligand | | 分子名称: | (E)-3-(2-(4-cyanostyryl)-4-oxoquinazolin-3(4H)-yl)benzoic acid, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Bouley, R, Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | 登録日 | 2013-12-21 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.947 Å) | | 主引用文献 | Discovery of Antibiotic (E)-3-(3-Carboxyphenyl)-2-(4-Cyanostyryl)Quinazolin-4(3H)-One.
J.Am.Chem.Soc., 137, 2015
|
|
4PB2
 
 | |
4PD7
 
 | | Structure of vcCNT bound to zebularine | | 分子名称: | DECYL-BETA-D-MALTOPYRANOSIDE, NupC family protein, SODIUM ION, ... | | 著者 | Johnson, Z.L, Lee, S.-Y. | | 登録日 | 2014-04-17 | | 公開日 | 2014-08-13 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.909 Å) | | 主引用文献 | Structural basis of nucleoside and nucleoside drug selectivity by concentrative nucleoside transporters.
Elife, 3, 2014
|
|
6PIB
 
 | | Structure of the Klebsiella pneumoniae LpxH-AZ1 complex | | 分子名称: | 1-[5-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | 著者 | Cho, J, Zhou, P. | | 登録日 | 2019-06-26 | | 公開日 | 2020-02-12 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7JMT
 
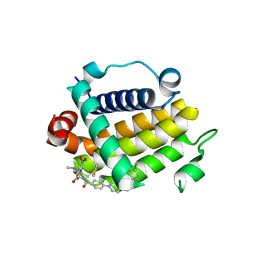 | | Crystal structure of schistosome BCL-2 bound to ABT-737 | | 分子名称: | 4-{4-[(4'-CHLOROBIPHENYL-2-YL)METHYL]PIPERAZIN-1-YL}-N-{[4-({(1R)-3-(DIMETHYLAMINO)-1-[(PHENYLTHIO)METHYL]PROPYL}AMINO)-3-NITROPHENYL]SULFONYL}BENZAMIDE, BCL-2 protein | | 著者 | Smith, N.A, Smith, B.J, Lee, E.F, Colman, P.M, Fairlie, W.D. | | 登録日 | 2020-08-02 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Optimization of Benzothiazole and Thiazole Hydrazones as Inhibitors of Schistosome BCL-2.
Acs Infect Dis., 7, 2021
|
|
5C22
 
 | | Crystal structure of Zn-bound HlyD from E. coli | | 分子名称: | Chromosomal hemolysin D, ZINC ION | | 著者 | Ha, N.C, Kim, J.S. | | 登録日 | 2015-06-15 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.302 Å) | | 主引用文献 | Crystal Structure of a Soluble Fragment of the Membrane Fusion Protein HlyD in a Type I Secretion System of Gram-Negative Bacteria
Structure, 24, 2016
|
|
5C21
 
 | |
6JN8
 
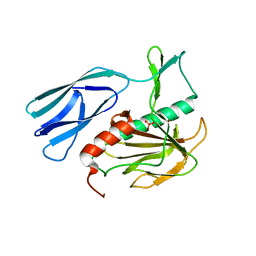 | | Structure of H216A mutant open form peptidoglycan peptidase | | 分子名称: | Peptidase M23, SULFATE ION, ZINC ION | | 著者 | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | 登録日 | 2019-03-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.106 Å) | | 主引用文献 | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
