5GTC
 
 | | Crystal structure of complex between DMAP-SH conjugated with a Kaposi's sarcoma herpesvirus LANA peptide (5-15) and nucleosome core particle | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Arimura, Y, Kato, D, Suto, H, Kurumizaka, H, Kawashima, S.A, Yamatsugu, K, Kanai, M. | | 登録日 | 2016-08-19 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Synthetic Posttranslational Modifications: Chemical Catalyst-Driven Regioselective Histone Acylation of Native Chromatin.
J. Am. Chem. Soc., 139, 2017
|
|
6FZ9
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A187F/L360F | | 分子名称: | CALCIUM ION, Lipase, ZINC ION | | 著者 | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | 登録日 | 2018-03-14 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.2463 Å) | | 主引用文献 | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZA
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A187F | | 分子名称: | CALCIUM ION, Lipase, ZINC ION | | 著者 | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | 登録日 | 2018-03-14 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZ8
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L184F/A187F | | 分子名称: | CALCIUM ION, Lipase, ZINC ION | | 著者 | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | 登録日 | 2018-03-14 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZD
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant L184F/A187F/L360F | | 分子名称: | CALCIUM ION, Lipase, ZINC ION | | 著者 | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | 登録日 | 2018-03-14 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZ1
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L360F | | 分子名称: | CALCIUM ION, Lipase, ZINC ION | | 著者 | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | 登録日 | 2018-03-13 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZC
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant L184F/L360F | | 分子名称: | CALCIUM ION, Lipase, ZINC ION | | 著者 | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | 登録日 | 2018-03-14 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZ7
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L184F | | 分子名称: | CALCIUM ION, Lipase, ZINC ION | | 著者 | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | 登録日 | 2018-03-14 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.736 Å) | | 主引用文献 | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
5BZ6
 
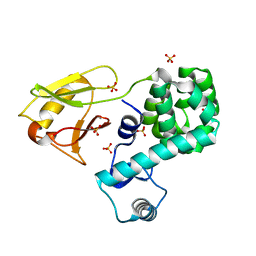 | | Crystal structure of the N-terminal domain single mutant (S92A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | 分子名称: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | 著者 | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | 登録日 | 2015-06-11 | | 公開日 | 2015-09-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
3W7B
 
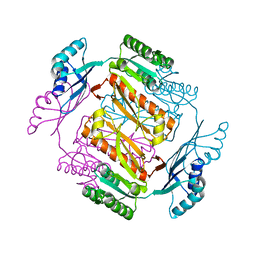 | | Crystal structure of formyltetrahydrofolate deformylase from Thermus thermophilus HB8 | | 分子名称: | Formyltetrahydrofolate deformylase | | 著者 | Sampei, G, Yanagida, Y, Ogata, N, Kusano, M, Terao, K, Kawai, H, Fukai, Y, Kanagawa, M, Inoue, Y, Baba, S, Kawai, G. | | 登録日 | 2013-02-28 | | 公開日 | 2014-01-08 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structures and reaction mechanisms of the two related enzymes, PurN and PurU
J.Biochem., 154, 2013
|
|
3WZN
 
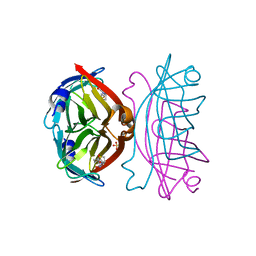 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with biotin at 1.3 A resolution | | 分子名称: | BIOTIN, SULFATE ION, Streptavidin | | 著者 | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | 登録日 | 2014-10-01 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3WY8
 
 | | Crystal Structure of Protease Anisep from Arthrobacter Nicotinovorans | | 分子名称: | Serine protease | | 著者 | Sone, T, Haraguchi, Y, Kuwahara, A, Ose, T, Takano, M, Abe, A, Tanaka, M, Tanaka, I, Asano, K. | | 登録日 | 2014-08-20 | | 公開日 | 2015-08-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural characterization reveals the keratinolytic activity of an arthrobacter nicotinovorans protease.
Protein Pept.Lett., 22, 2015
|
|
3WZP
 
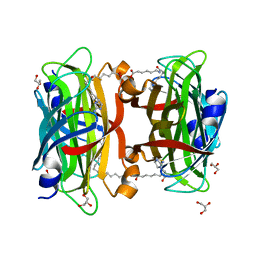 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.2 A resolution | | 分子名称: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, GLYCEROL, Streptavidin | | 著者 | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | 登録日 | 2014-10-01 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3WZQ
 
 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.7 A resolution | | 分子名称: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, HEXAETHYLENE GLYCOL, Streptavidin | | 著者 | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | 登録日 | 2014-10-01 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3WZO
 
 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with biotin long tail (BTNtail) at 1.5 A resolution | | 分子名称: | 6-({5-[(3aS,4S,5S,6aR)-5-oxido-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, CADMIUM ION, GLYCEROL, ... | | 著者 | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | 登録日 | 2014-10-01 | | 公開日 | 2015-02-18 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3X00
 
 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with bis iminobiotin long tail (Bis-IMNtail) at 1.3 A resolution | | 分子名称: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, ETHANE-1,2-DIAMINE, Streptavidin | | 著者 | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | 登録日 | 2014-10-09 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structure-based design and synthesis of a bivalent iminobiotin analog showing strong affinity toward a low immunogenic streptavidin mutant.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
3ZSK
 
 | | Crystal structure of Human Galectin-3 CRD with glycerol bound at 0.90 angstrom resolution | | 分子名称: | GALECTIN-3, GLYCEROL | | 著者 | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | 登録日 | 2011-06-28 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
3ZSJ
 
 | | Crystal structure of Human Galectin-3 CRD in complex with Lactose at 0.86 angstrom resolution | | 分子名称: | GALECTIN-3, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Saraboji, K, Hakansson, M, Diehl, C, Nilsson, U.J, Leffler, H, Akke, M, Logan, D.T. | | 登録日 | 2011-06-28 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (0.86 Å) | | 主引用文献 | The Carbohydrate-Binding Site in Galectin-3 is Pre-Organized to Recognize a Sugar-Like Framework of Oxygens: Ultra-High Resolution Structures and Water Dynamics.
Biochemistry, 51, 2012
|
|
4A6T
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in complex with PLP | | 分子名称: | OMEGA TRANSAMINASE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | 登録日 | 2011-11-08 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
4BFC
 
 | | Crystal structure of the C-terminal CMP-Kdo binding domain of WaaA from Acinetobacter baumannii | | 分子名称: | 3-DEOXY-D-MANNO-OCTULOSONIC-ACID TRANSFERASE, BETA-MERCAPTOETHANOL, SULFATE ION | | 著者 | Kimbung, Y.R, Hakansson, M, Logan, D, Wang, P.F, Schulz, M, Mamat, U, Woodard, R.W. | | 登録日 | 2013-03-18 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of the C-Terminal Cmp-Kdo Binding Domain of Waaa from Acinetobacter Baumannii
To be Published
|
|
4A6R
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in the apo form, crystallised from polyacrylic acid | | 分子名称: | OMEGA TRANSAMINASE, POLYACRYLIC ACID | | 著者 | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | 登録日 | 2011-11-08 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.349 Å) | | 主引用文献 | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
4A6U
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in the apo form, crystallised from PEG 3350 | | 分子名称: | OMEGA TRANSAMINASE, SODIUM ION, THIOCYANATE ION | | 著者 | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | 登録日 | 2011-11-08 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.687 Å) | | 主引用文献 | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
4A72
 
 | | Crystal structure of the omega transaminase from Chromobacterium violaceum in a mixture of apo and PLP-bound states | | 分子名称: | OMEGA TRANSAMINASE, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Logan, D.T, Hakansson, M, Yengo, K, Svedendahl Humble, M, Engelmark Cassimjee, K, Walse, B, Abedi, V, Federsel, H.-J, Berglund, P. | | 登録日 | 2011-11-10 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structures of the Chromobacterium Violaceum Omega-Transaminase Reveal Major Structural Rearrangements Upon Binding of Coenzyme Plp.
FEBS J., 279, 2012
|
|
2FAW
 
 | | crystal structure of papaya glutaminyl cyclase | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ZINC ION, ... | | 著者 | Wintjens, R, Belrhali, H, Clantin, B, Azarkan, M, Bompard, C, Baeyens-Volant, D, Looze, Y, Villeret, V. | | 登録日 | 2005-12-08 | | 公開日 | 2006-10-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of papaya glutaminyl cyclase, an archetype for plant and bacterial glutaminyl cyclases.
J.Mol.Biol., 357, 2006
|
|
4BJ0
 
 | | Xyloglucan binding module (CBM4-2 X2-L110F) in complex with branched xyloses | | 分子名称: | CALCIUM ION, XYLANASE, alpha-D-glucopyranose, ... | | 著者 | Schantz, L, Hakansson, M, Logan, D.T, Nordberg-Karlsson, E, Ohlin, M. | | 登録日 | 2013-04-15 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Carbohydrate Binding Module Recognition of Xyloglucan Defined by Polar Contacts with Branching Xyloses and Ch-Pi Interactions.
Proteins, 82, 2014
|
|
