1K3N
 
 | | NMR Structure of the FHA1 Domain of Rad53 in Complex with a Rad9-derived Phosphothreonine (at T155) Peptide | | 分子名称: | DNA repair protein Rad9, Protein Kinase SPK1 | | 著者 | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | 登録日 | 2001-10-03 | | 公開日 | 2001-12-05 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | 分子名称: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | 著者 | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | 登録日 | 2023-02-21 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8IGO
 
 | | Crystal structure of apo SARS-CoV-2 main protease | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | 登録日 | 2023-02-21 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
2N8U
 
 | |
1PHB
 
 | |
1J4P
 
 | | NMR STRUCTURE OF THE FHA1 DOMAIN OF RAD53 IN COMPLEX WITH A RAD9-DERIVED PHOSPHOTHREONINE (AT T155) PEPTIDE | | 分子名称: | DNA REPAIR PROTEIN RAD9, PROTEIN KINASE SPK1 | | 著者 | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | 登録日 | 2001-10-22 | | 公開日 | 2001-12-05 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
1PHA
 
 | |
2N8S
 
 | |
2N8T
 
 | |
2L16
 
 | |
2MN6
 
 | |
2YU2
 
 | |
2YU1
 
 | |
5XZR
 
 | | The atomic structure of SHP2 E76A mutant in complex with allosteric inhibitor 9b | | 分子名称: | 4-(3-phenylphenyl)-N-(2,2,6,6-tetramethylpiperidin-4-yl)-1,3-thiazol-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | 著者 | Li, D, Xie, J, Zhu, J, Liu, C. | | 登録日 | 2017-07-13 | | 公開日 | 2017-12-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Allosteric Inhibitors of SHP2 with Therapeutic Potential for Cancer Treatment.
J. Med. Chem., 60, 2017
|
|
4F66
 
 | | The crystal structure of 6-phospho-beta-glucosidase from Streptococcus mutans UA159 in complex with beta-D-glucose-6-phosphate. | | 分子名称: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-glucopyranose, FORMIC ACID, ... | | 著者 | Tan, K, Michalska, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-05-14 | | 公開日 | 2012-06-13 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.479 Å) | | 主引用文献 | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr. D Biol. Crystallogr., 69, 2013
|
|
8WYD
 
 | | Cryo-EM structure of DSR2-DSAD1 complex | | 分子名称: | Bacillus phage SPbeta DSAD1 protein, SIR2 family protein | | 著者 | Zhang, J.T, Jia, N, Liu, X.Y. | | 登録日 | 2023-10-30 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (2.56 Å) | | 主引用文献 | Structural basis for phage-mediated activation and repression of bacterial DSR2 anti-phage defense system.
Nat Commun, 15, 2024
|
|
8WYE
 
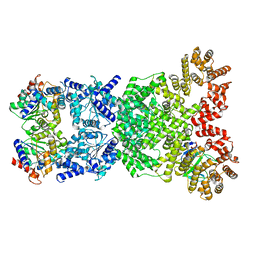 | |
8WY9
 
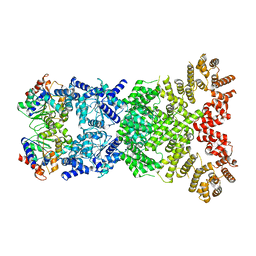 | |
3QOM
 
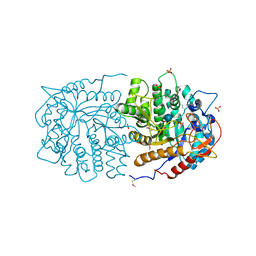 | | Crystal structure of 6-phospho-beta-glucosidase from Lactobacillus plantarum | | 分子名称: | 6-phospho-beta-glucosidase, ACETATE ION, PHOSPHATE ION, ... | | 著者 | Michalska, K, Hatzos-Skintges, C, Bearden, J, Kohler, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-02-10 | | 公開日 | 2011-03-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.498 Å) | | 主引用文献 | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8WYF
 
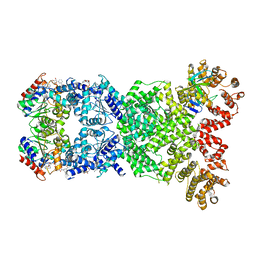 | |
8XLQ
 
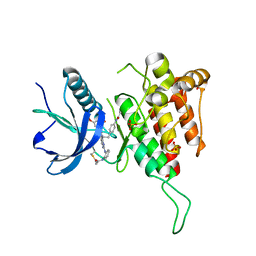 | |
8XLO
 
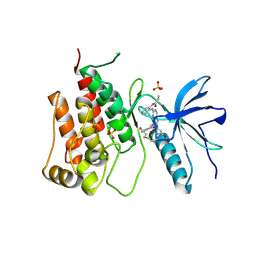 | |
4FMX
 
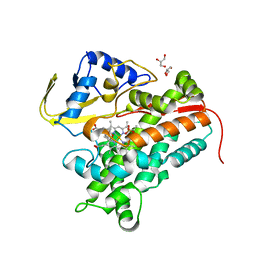 | | Crystal Structure of Substrate-Bound P450cin | | 分子名称: | 1,3,3-TRIMETHYL-2-OXABICYCLO[2.2.2]OCTANE, GLYCEROL, P450cin, ... | | 著者 | Madrona, Y, Tripathi, S.M, Huiying, L, Poulos, T.L. | | 登録日 | 2012-06-18 | | 公開日 | 2012-07-25 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.554 Å) | | 主引用文献 | Crystal structures of substrate-free and nitrosyl cytochrome p450cin: implications for o(2) activation.
Biochemistry, 51, 2012
|
|
4GZE
 
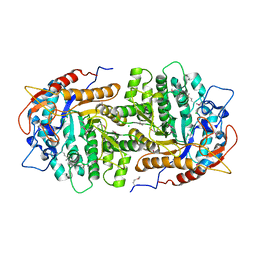 | | Crystal structure of 6-phospho-beta-glucosidase from Lactobacillus plantarum (apo form) | | 分子名称: | 6-phospho-beta-glucosidase, CHLORIDE ION, GLYCEROL | | 著者 | Michalska, K, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2012-09-06 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | GH1-family 6-P-beta-glucosidases from human microbiome lactic acid bacteria.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1MOC
 
 | |
