2CTZ
 
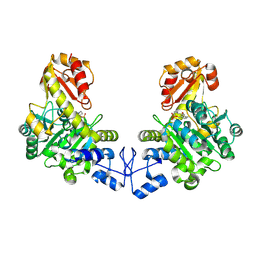 | | Crystal structure of o-acetyl homoserine sulfhydrylase from Thermus thermophilus HB8 | | 分子名称: | O-acetyl-L-homoserine sulfhydrylase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Imagawa, T, Kousumi, Y, Tsuge, H, Utsunomiya, H, Ebihara, A, Nakagawa, N, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2005-05-24 | | 公開日 | 2005-11-24 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of o-acetyl homoserine sulfhydrylase from Thermus thermophilus HB8
To be Published
|
|
6JZT
 
 | |
6K3I
 
 | |
6JZR
 
 | |
6KMF
 
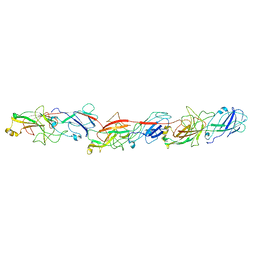 | | FimA type V pilus from P.gingivalis | | 分子名称: | Major fimbrium subunit FimA type-1 | | 著者 | Shibata, S, Shoji, M, Matsunami, H, Matthews, M, Imada, K, Nakayama, K, Wolf, M. | | 登録日 | 2019-07-31 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structure of polymerized type V pilin reveals assembly mechanism involving protease-mediated strand exchange.
Nat Microbiol, 5, 2020
|
|
6JF2
 
 | |
7VFD
 
 | |
7VFE
 
 | |
7VFG
 
 | |
7VFH
 
 | |
7VFF
 
 | |
5Z9W
 
 | | Ebola virus nucleoprotein-RNA complex | | 分子名称: | Ebolavirus nucleoprotein (residues 19-406), RNA (6-MER) | | 著者 | Sugita, Y, Matsunami, H, Kawaoka, Y, Noda, T, Wolf, M. | | 登録日 | 2018-02-05 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 angstrom resolution.
Nature, 563, 2018
|
|
2D4Y
 
 | |
2D4X
 
 | |
8HTU
 
 | | Cryo-EM structure of PpPSI-L | | 分子名称: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | 著者 | Li, M, Pan, X.W, Sun, H.Y. | | 登録日 | 2022-12-21 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Structural insights into the assembly and energy transfer of the Lhcb9-dependent photosystem I from moss Physcomitrium patens.
Nat.Plants, 9, 2023
|
|
5B6I
 
 | |
8JUB
 
 | | Crystal structure of glutaminase C in complex with compound 27 | | 分子名称: | 3-[2-oxidanylidene-2-[[5-[[(3R)-1-pyridazin-3-ylpyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]amino]ethyl]benzoic acid, Glutaminase kidney isoform, mitochondrial | | 著者 | Wang, X, Hanyu, S, Tingting, D. | | 登録日 | 2023-06-26 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Targeting the Subpocket Enables the Discovery of Thiadiazole-Pyridazine Derivatives as Glutaminase C Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
8JUE
 
 | | Crystal structure of glutaminase C in complex with compound 11 | | 分子名称: | 2-(3-phenoxyphenyl)-N-[5-[[(3R)-1-pyridazin-3-ylpyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial | | 著者 | Wang, X, Hanyu, S, Tingting, D. | | 登録日 | 2023-06-26 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Targeting the Subpocket Enables the Discovery of Thiadiazole-Pyridazine Derivatives as Glutaminase C Inhibitors.
Acs Med.Chem.Lett., 14, 2023
|
|
2JK7
 
 | | XIAP BIR3 bound to a Smac Mimetic | | 分子名称: | (3S,6S,7Z,10AS)-N-(DIPHENYLMETHYL)-6-{[(2S)-2-(METHYLIDENEAMINO)BUTANOYL]AMINO}-5-OXO-1,2,3,5,6,9,10,10A-OCTAHYDROPYRROLO[1,2-A]AZOCINE-3-CARBOXAMIDE, BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 4, ZINC ION | | 著者 | Saito, N.G, Meagher, J.L, Stuckey, J.A. | | 登録日 | 2008-08-21 | | 公開日 | 2008-11-04 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Structure-Based Design, Synthesis, Evaluation, and Crystallographic Studies of Conformationally Constrained Smac Mimetics as Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (Xiap).
J.Med.Chem., 51, 2008
|
|
5EC9
 
 | | Retinoic acid receptor alpha in complex with chiral dihydrobenzofuran benzoic acid 9a and a fragment of the coactivator TIF2 | | 分子名称: | 4-[(11S,15R)-4,4,7,7-Tetramethyl-16-oxatetracyclo[8.6.0.03,8.011,15]hexadeca-1(10),2,8-trien-11-yl]benzoic acid, LYS-HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP, Retinoic acid receptor RXR-alpha | | 著者 | Leysen, S, Ottmann, C, Schafer, A, Scheepstra, M, Brunsveld, L, Sunden, R, Ma, J.N, Burnstein, E.S, Olsson, R. | | 登録日 | 2015-10-20 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Chiral Dihydrobenzofuran Acids Show Potent Retinoid X Receptor-Nuclear Receptor Related 1 Protein Dimer Activation.
J.Med.Chem., 59, 2016
|
|
5QTT
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(3R,7S)-7-{[5-amino-1-(3-chloro-2-fluorophenyl)-1H-pyrazole-4-carbonyl]amino}-3-methyl-2-oxo-2,3,4,5,6,7-hexahydro-1H-12,8-(metheno)-1,9-benzodiazacyclotetradecin-15-yl]carbamate | | 分子名称: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | 著者 | Sheriff, S. | | 登録日 | 2019-10-16 | | 公開日 | 2019-12-25 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Potent, Orally Bioavailable and Efficacious Macrocyclic Inhibitors of Factor XIa. Discovery of Pyridine-Based Macrocycles Possessing Phenylazole Carboxamide P1 Groups.
J.Med.Chem., 63, 2019
|
|
5QTU
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(3R,7S)-7-{[1-(3-chloro-2-fluorophenyl)-5-methyl-1H-imidazole-4-carbonyl]amino}-3-methyl-2-oxo-2,3,4,5,6,7-hexahydro-1H-12,8-(metheno)-1,9-benzodiazacyclotetradecin-15-yl]carbamate | | 分子名称: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | 著者 | Sheriff, S. | | 登録日 | 2019-10-16 | | 公開日 | 2019-12-25 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Potent, Orally Bioavailable and Efficacious Macrocyclic Inhibitors of Factor XIa. Discovery of Pyridine-Based Macrocycles Possessing Phenylazole Carboxamide P1 Groups.
J.Med.Chem., 63, 2019
|
|
6XLI
 
 | | CRYSTAL STRUCTURE OF ANTI-TAU ANTIBODY PT3 Fab+pT212/pT217-TAU PEPTIDE | | 分子名称: | GLYCEROL, PT3 Fab Heavy Chain, PT3 Fab Light Chain, ... | | 著者 | Malia, T.J, Teplyakov, A, Luo, J. | | 登録日 | 2020-06-28 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery and Functional Characterization of hPT3, a Humanized Anti-Phospho Tau Selective Monoclonal Antibody.
J Alzheimers Dis, 77, 2020
|
|
8COI
 
 | | Human adenovirus-derived synthetic ADDobody binder | | 分子名称: | ADDobody | | 著者 | Buzas, D, Toelzer, C, Gupta, K, Berger-Schaffitzel, C, Berger, I. | | 登録日 | 2023-02-28 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.17 Å) | | 主引用文献 | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
5QTW
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl (2R,7S)-7-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-14-[(methoxycarbonyl)amino]-2,3,4,5,6,7-hexahydro-1H-8,11-epimino-1,9-benzodiazacyclotridecine-2-carboxylate | | 分子名称: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | 著者 | Sheriff, S. | | 登録日 | 2019-11-13 | | 公開日 | 2020-01-29 | | 最終更新日 | 2021-05-12 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Orally bioavailable amine-linked macrocyclic inhibitors of factor XIa.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
