5MHT
 
 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH HEMIMETHYLATED DNA AND ADOHCY | | 分子名称: | DNA (5'-D(*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3'), PROTEIN (HHAI METHYLTRANSFERASE), ... | | 著者 | Cheng, X. | | 登録日 | 1996-10-22 | | 公開日 | 1997-07-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A structural basis for the preferential binding of hemimethylated DNA by HhaI DNA methyltransferase.
J.Mol.Biol., 263, 1996
|
|
7F8S
 
 | | Pennisetum glaucum (Pearl millet) dehydroascorbate reductase (DHAR) with catalytic cysteine (Cy20) in sulphenic and sulfinic acid forms. | | 分子名称: | Dehydroascorbate reductase, SULFATE ION | | 著者 | Das, B.K, Kumar, A, Sreeshma, N.S, Arockiasamy, A. | | 登録日 | 2021-07-02 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Comparative kinetic analysis of ascorbate (Vitamin-C) recycling dehydroascorbate reductases from plants and humans.
Biochem.Biophys.Res.Commun., 591, 2021
|
|
2MI1
 
 | |
1KBS
 
 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 1 STRUCTURE | | 分子名称: | CTX IV | | 著者 | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | 登録日 | 1996-07-22 | | 公開日 | 1997-07-23 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
1KBT
 
 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 12 STRUCTURES | | 分子名称: | CTX IV | | 著者 | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | 登録日 | 1996-07-22 | | 公開日 | 1997-07-23 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
6LW8
 
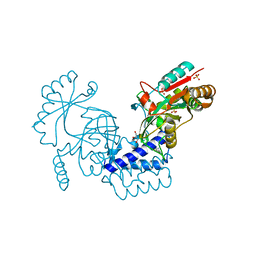 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | 分子名称: | (4R)-4-(4-fluorophenyl)-4,5,6,7-tetrahydro-1H-imidazo[4,5-c]pyridine, DNA ligase A, GLYCEROL, ... | | 著者 | Ramachandran, R, Afsar, M, Shukla, A. | | 登録日 | 2020-02-07 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Structure based identification of first-in-class fragment inhibitors that target the NMN pocket of M. tuberculosis NAD + -dependent DNA ligase A.
J.Struct.Biol., 213, 2021
|
|
7U0X
 
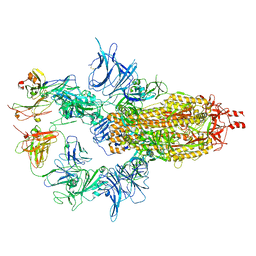 | |
7U0Q
 
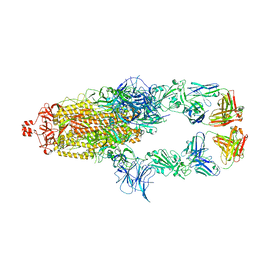 | |
1R5O
 
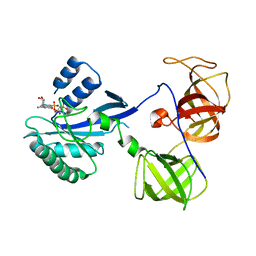 | | crystal structure analysis of sup35 complexed with GMPPNP | | 分子名称: | Eukaryotic peptide chain release factor GTP-binding subunit, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Kong, C, Song, H. | | 登録日 | 2003-10-11 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
8EUA
 
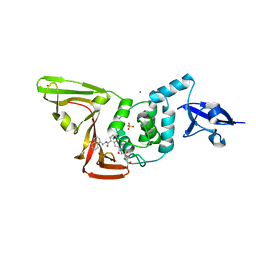 | | Structure of SARS-CoV2 PLpro bound to a covalent inhibitor | | 分子名称: | Papain-like protease nsp3, SULFATE ION, ZINC ION, ... | | 著者 | Mathews, I.I, Pokhrel, S, Wakatsuki, S. | | 登録日 | 2022-10-18 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2.
Nat Commun, 14, 2023
|
|
2PQM
 
 | |
1R5B
 
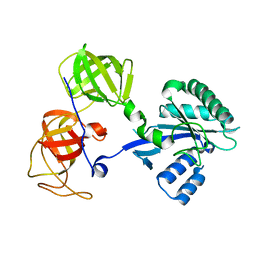 | | Crystal structure analysis of sup35 | | 分子名称: | Eukaryotic peptide chain release factor GTP-binding subunit | | 著者 | Kong, C, Song, H. | | 登録日 | 2003-10-10 | | 公開日 | 2004-05-25 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
1R5N
 
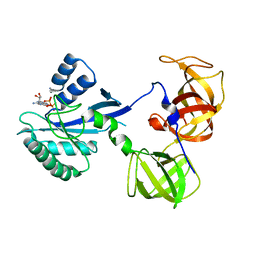 | | Crystal Structure Analysis of sup35 complexed with GDP | | 分子名称: | Eukaryotic peptide chain release factor GTP-binding subunit, GUANOSINE-5'-DIPHOSPHATE | | 著者 | Kong, C, Song, H. | | 登録日 | 2003-10-10 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe
Mol.Cell, 14, 2004
|
|
8A8N
 
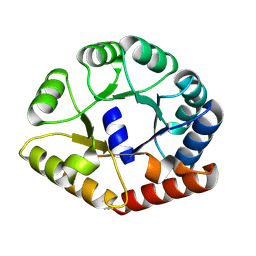 | |
5YO2
 
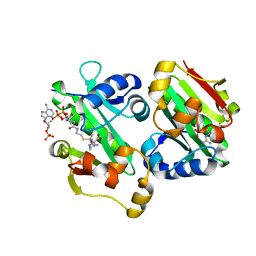 | | The crystal structure of Rv2747 from Mycobacterium tuberculosis in complex with Acetyl CoA and L-Arginine | | 分子名称: | ACETYL COENZYME *A, ARGININE, Amino-acid acetyltransferase | | 著者 | Singh, E, Tiruttani Subhramanyam, U.K, Pal, R.K, Srinivasan, A, Gourinath, S, Das, U. | | 登録日 | 2017-10-26 | | 公開日 | 2018-11-07 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.997 Å) | | 主引用文献 | Structural insights into the substrate binding mechanism of novel ArgA from Mycobacterium tuberculosis
Int. J. Biol. Macromol., 125, 2019
|
|
6D2P
 
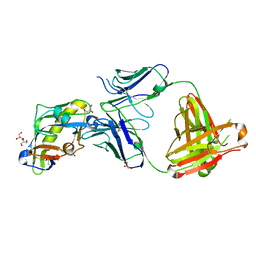 | |
6ADD
 
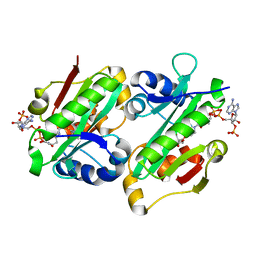 | | The crystal structure of Rv2747 from Mycobacterium tuberculosis in complex with CoA and NLQ | | 分子名称: | Amino-acid acetyltransferase, COENZYME A, N~2~-ACETYL-L-GLUTAMINE | | 著者 | Das, U, Singh, E, Pal, R.K, Tiruttani Subhramanyam, U.K, Gourinath, S, Srinivasan, A. | | 登録日 | 2018-07-31 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | Structural insights into the substrate binding mechanism of novel ArgA from Mycobacterium tuberculosis
Int. J. Biol. Macromol., 125, 2019
|
|
2SAS
 
 | |
1UBI
 
 | | SYNTHETIC STRUCTURAL AND BIOLOGICAL STUDIES OF THE UBIQUITIN SYSTEM. PART 1 | | 分子名称: | UBIQUITIN | | 著者 | Alexeev, D, Bury, S.M, Turner, M.A, Ogunjobi, O.M, Muir, T.W, Ramage, R, Sawyer, L. | | 登録日 | 1994-02-03 | | 公開日 | 1994-05-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Synthetic, structural and biological studies of the ubiquitin system: the total chemical synthesis of ubiquitin.
Biochem.J., 299, 1994
|
|
5ZAU
 
 | |
1VE3
 
 | | Crystal structure of PH0226 protein from Pyrococcus horikoshii OT3 | | 分子名称: | S-ADENOSYLMETHIONINE, hypothetical protein PH0226 | | 著者 | Lokanath, N.K, Yamamoto, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-03-26 | | 公開日 | 2005-05-24 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of SAM-dependent methyltransferase from Pyrococcus horikoshii.
Acta Crystallogr.,Sect.F, 73, 2017
|
|
5YB0
 
 | |
5YD2
 
 | |
5YII
 
 | |
3MHT
 
 | | TERNARY STRUCTURE OF HHAI METHYLTRANSFERASE WITH UNMODIFIED DNA AND ADOHCY | | 分子名称: | DNA (5'-D(*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*TP*AP*GP*CP*GP*CP*TP*AP*TP*C)-3'), PROTEIN (HHAI METHYLTRANSFERASE (E.C.2.1.1.73)), ... | | 著者 | O'Gara, M, Klimasauskas, S, Roberts, R.J, Cheng, X. | | 登録日 | 1996-07-24 | | 公開日 | 1997-01-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaL methyltransferase-DNA-AdoHcy complexes.
J.Mol.Biol., 261, 1996
|
|
