7KCV
 
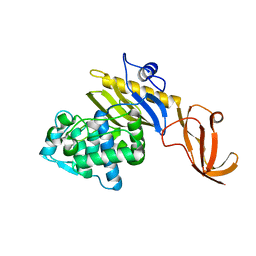 | |
7KCY
 
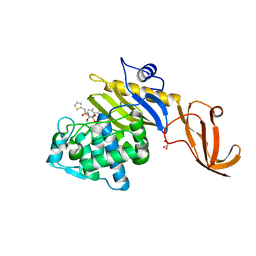 | | Crystal structure of S. aureus penicillin-binding protein 4 (PBP4) with cefoxitin | | 分子名称: | (2R)-2-{(1S)-1-methoxy-2-oxo-1-[(thiophen-2-ylacetyl)amino]ethyl}-5-methylidene-5,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, Penicillin-binding protein 4, ... | | 著者 | Alexander, J.A.N, Strynadka, N.C.J. | | 登録日 | 2020-10-07 | | 公開日 | 2021-06-30 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | PBP4-mediated beta-lactam resistance among clinical strains of Staphylococcus aureus.
J.Antimicrob.Chemother., 76, 2021
|
|
5DU8
 
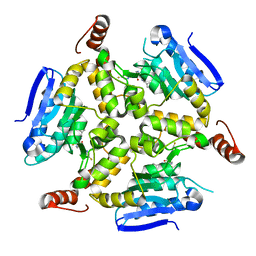 | | Crystal structure of M. tuberculosis EchA6 bound to GSK572A | | 分子名称: | (5R,7S)-5-(4-ethylphenyl)-N-[(5-fluoropyridin-2-yl)methyl]-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | 著者 | Cox, J.A.G, Besra, G.S, Futterer, K. | | 登録日 | 2015-09-18 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
7LHX
 
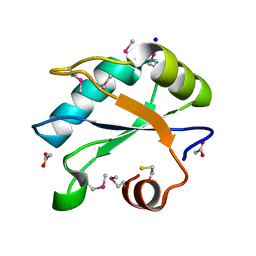 | | Human U1A protein with F37M and F77M mutations for improved phasing | | 分子名称: | ACETATE ION, BETA-MERCAPTOETHANOL, SODIUM ION, ... | | 著者 | Jenkins, J.L, Lippa, G.M, Wedekind, J.E. | | 登録日 | 2021-01-26 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Affinity and Structural Analysis of the U1A RNA Recognition Motif with Engineered Methionines to Improve Experimental Phasing
Crystals, 11, 2021
|
|
5DTP
 
 | |
5DU4
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK366A | | 分子名称: | (5R,7S)-5-(4-ethylphenyl)-N-(4-methoxybenzyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | 著者 | Cox, J.A.G, Besra, G.S, Futterer, K. | | 登録日 | 2015-09-18 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.702 Å) | | 主引用文献 | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5DU6
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK059A. | | 分子名称: | (5R,7R)-5-(4-ethylphenyl)-N-(4-fluorobenzyl)-7-methyl-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | 著者 | Cox, J.A.G, Besra, G.S, Futterer, K. | | 登録日 | 2015-09-18 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.61 Å) | | 主引用文献 | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5DTW
 
 | |
5DUF
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK729A | | 分子名称: | (5R,7S)-5-(4-ethylphenyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxylic acid, Probable enoyl-CoA hydratase echA6 | | 著者 | Cox, J.A.G, Besra, G.S, Futterer, K. | | 登録日 | 2015-09-18 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
5DUC
 
 | | Crystal structure of M. tuberculosis EchA6 bound to ligand GSK951A | | 分子名称: | (5R,7S)-N-(1,3-benzodioxol-5-ylmethyl)-5-(4-ethylphenyl)-7-(trifluoromethyl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, Probable enoyl-CoA hydratase echA6 | | 著者 | Cox, J.A.G, Besra, G.S, Futterer, K. | | 登録日 | 2015-09-18 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.704 Å) | | 主引用文献 | THPP target assignment reveals EchA6 as an essential fatty acid shuttle in mycobacteria.
Nat Microbiol, 1, 2016
|
|
1PDX
 
 | | PUTIDAREDOXIN | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, PROTEIN (PUTIDAREDOXIN) | | 著者 | Pochapsky, T.C, Jain, N.U, Kuti, M, Lyons, T.A, Heymont, J. | | 登録日 | 1999-02-15 | | 公開日 | 1999-05-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A refined model for the solution structure of oxidized putidaredoxin.
Biochemistry, 38, 1999
|
|
1PUT
 
 | | AN NMR-DERIVED MODEL FOR THE SOLUTION STRUCTURE OF OXIDIZED PUTIDAREDOXIN, A 2FE, 2-S FERREDOXIN FROM PSEUDOMONAS | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, PUTIDAREDOXIN | | 著者 | Pochapsky, T.C, Ye, X.M, Ratnaswamy, G, Lyons, T.A. | | 登録日 | 1994-07-09 | | 公開日 | 1994-09-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An NMR-derived model for the solution structure of oxidized putidaredoxin, a 2-Fe, 2-S ferredoxin from Pseudomonas.
Biochemistry, 33, 1994
|
|
2HJI
 
 | | Structural model for the Fe-containing isoform of acireductone dioxygenase | | 分子名称: | E-2/E-2' protein, FE (II) ION | | 著者 | Pochapsky, T.C, Ju, T, Maroney, M.J, Chai, S.C. | | 登録日 | 2006-06-30 | | 公開日 | 2006-10-24 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | One Protein, Two Enzymes Revisited: A Structural Entropy Switch Interconverts the Two Isoforms of Acireductone Dioxygenase
J.Mol.Biol., 363, 2006
|
|
4KBO
 
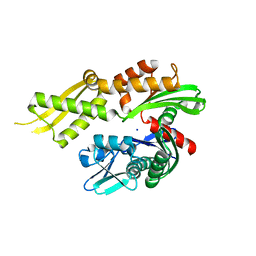 | | Crystal structure of the human Mortalin (GRP75) ATPase domain in the apo form | | 分子名称: | SODIUM ION, Stress-70 protein, mitochondrial | | 著者 | Amick, J, Page, R.C, Nix, J.C, Misra, S. | | 登録日 | 2013-04-23 | | 公開日 | 2014-04-02 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structure of the nucleotide-binding domain of mortalin, the mitochondrial Hsp70 chaperone.
Protein Sci., 23, 2014
|
|
2MTP
 
 | |
5V3F
 
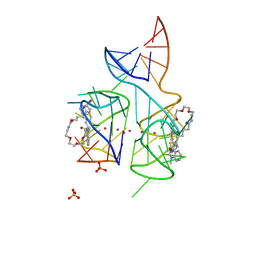 | | Co-crystal structure of the fluorogenic RNA Mango | | 分子名称: | 4-{[(2S)-3-{2,16-dioxo-20-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-6,9,12-trioxa-3,15-diazaicosan-1-yl}-2,3-dihydro-1,3-benzothiazol-2-yl]methyl}-1-methylquinolin-1-ium, PHOSPHATE ION, POTASSIUM ION, ... | | 著者 | Trachman, R.J, Ferre-D'Amare, A.R. | | 登録日 | 2017-03-07 | | 公開日 | 2017-05-24 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for high-affinity fluorophore binding and activation by RNA Mango.
Nat. Chem. Biol., 13, 2017
|
|
