1IPK
 
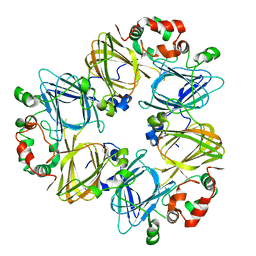 | | CRYSTAL STRUCTURES OF RECOMBINANT AND NATIVE SOYBEAN BETA-CONGLYCININ BETA HOMOTRIMERS | | 分子名称: | BETA-CONGLYCININ, BETA CHAIN | | 著者 | Maruyama, N, Adachi, M, Takahashi, K, Yagasaki, K, Kohno, M, Takenaka, Y, Okuda, E, Nakagawa, S, Mikami, B, Utsumi, S. | | 登録日 | 2001-05-16 | | 公開日 | 2002-05-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of recombinant and native soybean beta-conglycinin beta homotrimers.
Eur.J.Biochem., 268, 2001
|
|
7CII
 
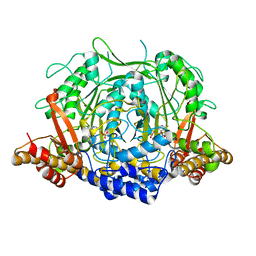 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with L- methionine methyl ester (external aldimine form). | | 分子名称: | L-methionine decarboxylase, methyl (2S)-2-[(E)-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]-4-methylsulfanyl-butanoate | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CIM
 
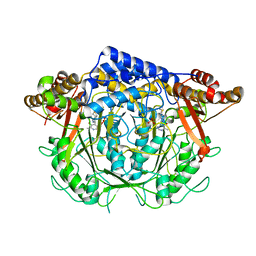 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with 3-methlythiopropylamine (geminal diamine form). | | 分子名称: | L-methionine decarboxylase, [6-methyl-4-[(3-methylsulfanylpropylamino)methyl]-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CIJ
 
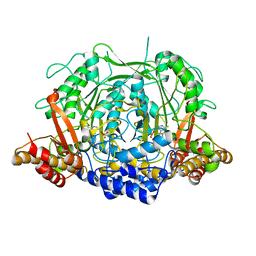 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 in complexed with 3-methlythiopropylamine (external aldimine form). | | 分子名称: | L-methionine decarboxylase, [6-methyl-4-[(E)-3-methylsulfanylpropyliminomethyl]-5-oxidanyl-pyridin-3-yl]methyl dihydrogen phosphate | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CIG
 
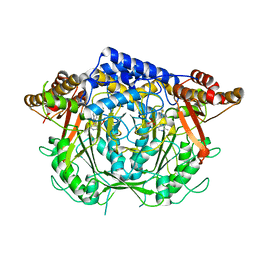 | | Crystal structure of L-methionine decarboxylase Q64A mutant from Streptomyces sp.590 in complexed with L- methionine methyl ester (geminal diamine form). | | 分子名称: | L-methionine decarboxylase, methyl (2S)-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-methylsulfanyl-butanoate | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
7CIF
 
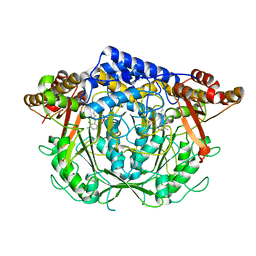 | | Crystal structure of L-methionine decarboxylase from Streptomyces sp.590 (internal aldimine form). | | 分子名称: | L-methionine decarboxylase | | 著者 | Okawa, A, Shiba, T, Hayashi, M, Onoue, Y, Murota, M, Sato, D, Inagaki, J, Tamura, T, Harada, S, Inagaki, K. | | 登録日 | 2020-07-07 | | 公開日 | 2021-01-27 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for substrate specificity of l-methionine decarboxylase.
Protein Sci., 30, 2021
|
|
3GCC
 
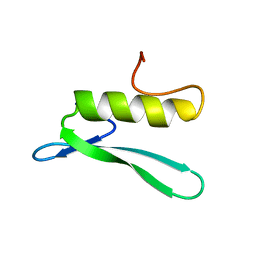 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, 46 STRUCTURES | | 分子名称: | ATERF1 | | 著者 | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | 登録日 | 1998-03-13 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
2GCC
 
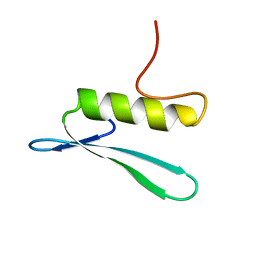 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | | 分子名称: | ATERF1 | | 著者 | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | 登録日 | 1998-03-13 | | 公開日 | 1999-03-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
7F2P
 
 | | The head structure of Helicobacter pylori bacteriophage KHP40 | | 分子名称: | Cement protein gp16, KHP40 MCP | | 著者 | Kamiya, R, Uchiyama, J, Matsuzaki, S, Murata, K, Iwasaki, K, Miyazaki, N. | | 登録日 | 2021-06-13 | | 公開日 | 2021-10-27 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Acid-stable capsid structure of Helicobacter pylori bacteriophage KHP30 by single-particle cryoelectron microscopy.
Structure, 30, 2022
|
|
6KLW
 
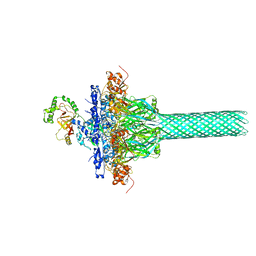 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with long stem | | 分子名称: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | 著者 | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | 登録日 | 2019-07-30 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2D4V
 
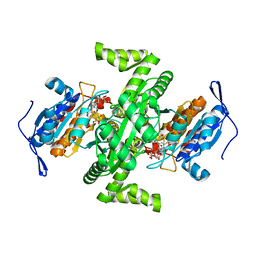 | | Crystal structure of NAD dependent isocitrate dehydrogenase from Acidithiobacillus thiooxidans | | 分子名称: | CITRATE ANION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, isocitrate dehydrogenase | | 著者 | Imada, K, Tamura, T, Namba, K, Inagaki, K. | | 登録日 | 2005-10-24 | | 公開日 | 2006-11-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and quantum chemical analysis of NAD+-dependent isocitrate dehydrogenase: hydride transfer and co-factor specificity
Proteins, 70, 2008
|
|
6KLX
 
 | | Pore structure of Iota toxin binding component (Ib) | | 分子名称: | CALCIUM ION, Iota toxin component Ib | | 著者 | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | 登録日 | 2019-07-30 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1NZ8
 
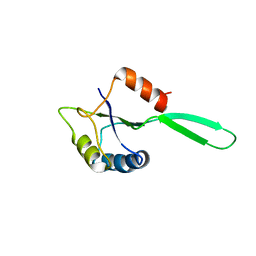 | | Solution Structure of the N-utilization substance G (NusG) N-terminal (NGN) domain from Thermus thermophilus | | 分子名称: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | 著者 | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-02-17 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
3VKF
 
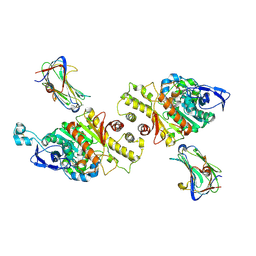 | | Crystal Structure of Neurexin 1beta/Neuroligin 1 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-1-beta, ... | | 著者 | Tanaka, H, Miyazaki, N, Nogi, T, Iwasaki, K, Takagi, J. | | 登録日 | 2011-11-15 | | 公開日 | 2012-08-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin.
Cell Rep, 2, 2012
|
|
1NZ9
 
 | | Solution Structure of the N-utilization substance G (NusG) C-terminal (NGC) domain from Thermus thermophilus | | 分子名称: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | 著者 | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-02-17 | | 公開日 | 2004-04-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
5XBR
 
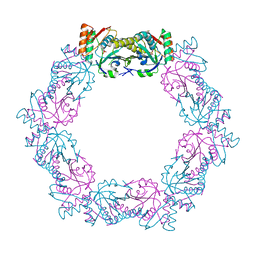 | |
5TT4
 
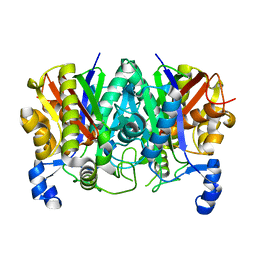 | | Determining the Molecular Basis For Starter Unit Selection During Daunorubicin Biosynthesis | | 分子名称: | Daunorubicin-doxorubicin polyketide synthase | | 著者 | Jackson, D.R, Valentic, T.R, Patel, A, Tsai, S.C, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | 登録日 | 2016-11-01 | | 公開日 | 2016-11-23 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Determining the Molecular Basis For Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
5XBQ
 
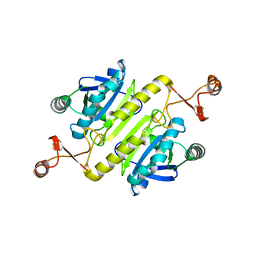 | |
6KLO
 
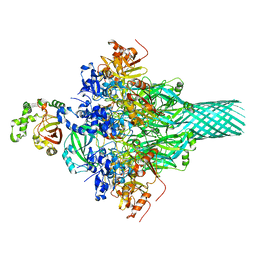 | | Complex structure of Iota toxin enzymatic component (Ia) and binding component (Ib) pore with short stem | | 分子名称: | CALCIUM ION, Iota toxin component Ia, Iota toxin component Ib | | 著者 | Yoshida, T, Yamada, T, Kawamoto, A, Mitsuoka, K, Iwasaki, K, Tsuge, H. | | 登録日 | 2019-07-30 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-EM structures reveal translocational unfolding in the clostridial binary iota toxin complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2RT4
 
 | |
2D4W
 
 | |
3A8U
 
 | |
3W3E
 
 | |
4XSA
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | 分子名称: | Daunorubicin-doxorubicin polyketide synthase | | 著者 | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | 登録日 | 2015-01-22 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.204 Å) | | 主引用文献 | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
4XS7
 
 | | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis | | 分子名称: | Daunorubicin-doxorubicin polyketide synthase | | 著者 | Jackson, D.R, Valentic, T.R, Tsai, S.C, Patel, A, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | 登録日 | 2015-01-22 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Determining the Molecular Basis for Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
