8UEO
 
 | | In-situ complex I (Active-Apo) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | 著者 | Zheng, W, Zhu, J, Zhang, K. | | 登録日 | 2023-10-02 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
8UGG
 
 | | In-situ complex III, state IV | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | 著者 | Zheng, W, Zhang, K, Zhu, J. | | 登録日 | 2023-10-05 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
8UEW
 
 | | In-situ complex I, Deactive class05 | | 分子名称: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | 著者 | Zheng, W, Zhu, J, Zhang, K. | | 登録日 | 2023-10-02 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
8UGP
 
 | | In-situ structure of typeA supercomplex in respiratory chain ( local refined map focused on CI iron-sulfur cluster regions ) | | 分子名称: | IRON/SULFUR CLUSTER, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6, ... | | 著者 | Zheng, W, Zhang, K, Zhu, J. | | 登録日 | 2023-10-05 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2 Å) | | 主引用文献 | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
8UES
 
 | | In-situ complex I, Deactive class01 | | 分子名称: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | 著者 | Zheng, W, Zhu, J, Zhang, K. | | 登録日 | 2023-10-02 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
8UER
 
 | | In-situ complex I with Q10 (State-gamma) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | 著者 | Zheng, W, Zhu, J, Zhang, K. | | 登録日 | 2023-10-02 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
8UGJ
 
 | | In-situ structure of typeB supercomplex in respiratory chain (composite) | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | 著者 | Zheng, W, Zhang, K, Zhu, J. | | 登録日 | 2023-10-05 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.3 Å) | | 主引用文献 | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
8UGK
 
 | | High resolution in-situ structure of complex III in respiratory supercomplex (composite) | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | 著者 | Zheng, W, Zhu, J, Zhang, K. | | 登録日 | 2023-10-05 | | 公開日 | 2024-06-19 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | High-resolution in situ structures of mammalian respiratory supercomplexes.
Nature, 631, 2024
|
|
3TK9
 
 | | Crystal structure of human granzyme H | | 分子名称: | Granzyme H, SULFATE ION | | 著者 | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | 登録日 | 2011-08-25 | | 公開日 | 2011-12-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
3QQ2
 
 | |
3TJU
 
 | | Crystal structure of human granzyme H with an inhibitor | | 分子名称: | Ac-PTSY-CMK inhibitor, Granzyme H, SULFATE ION | | 著者 | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | 登録日 | 2011-08-25 | | 公開日 | 2011-12-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
3TJV
 
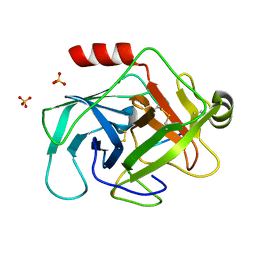 | | Crystal structure of human granzyme H with a peptidyl substrate | | 分子名称: | Granzyme H, PTSYAGDDSG, SULFATE ION | | 著者 | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | 登録日 | 2011-08-25 | | 公開日 | 2011-12-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
8UYJ
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | 分子名称: | BtCoV-HKU5 5' proximal stem-loop 5, conformation 4 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYS
 
 | | SARS-CoV-2 5' proximal stem-loop 5 | | 分子名称: | SARS-CoV-2 RNA SL5 domain. | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-14 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYL
 
 | | MERS 5' proximal stem-loop 5, conformation 2 | | 分子名称: | MERS 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYM
 
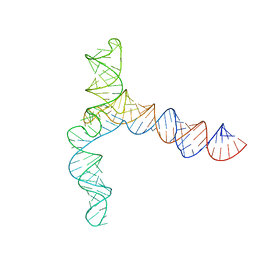 | | MERS 5' proximal stem-loop 5, conformation 3 | | 分子名称: | MERS 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYK
 
 | | MERS 5' proximal stem-loop 5, conformation 1 | | 分子名称: | MERS 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.9 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYE
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 1 | | 分子名称: | BtCoV-HKU5 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (5.9 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYG
 
 | | BtCoV-HKU5 5' proximal stem-loop 5, conformation 2 | | 分子名称: | RNA (135-MER) | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UYP
 
 | | SARS-CoV-1 5' proximal stem-loop 5 | | 分子名称: | SARS-CoV-1 5' proximal stem-loop 5 | | 著者 | Kretsch, R.C, Xu, L, Zheludev, I.N, Zhou, X, Huang, R, Nye, G, Li, S, Zhang, K, Chiu, W, Das, R. | | 登録日 | 2023-11-13 | | 公開日 | 2023-12-20 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Tertiary folds of the SL5 RNA from the 5' proximal region of SARS-CoV-2 and related coronaviruses.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
4F1P
 
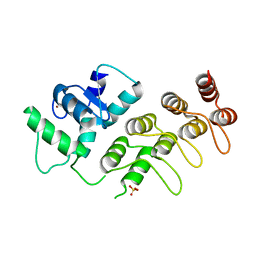 | | Crystal Structure of mutant S554D for ArfGAP and ANK repeat domain of ACAP1 | | 分子名称: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1, SULFATE ION, ... | | 著者 | Sun, F, Pang, X, Zhang, K, Ma, J, Zhou, Q. | | 登録日 | 2012-05-07 | | 公開日 | 2012-07-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Mechanistic insights into regulated cargo binding by ACAP1 protein
J.Biol.Chem., 287, 2012
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | 分子名称: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | 著者 | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | 登録日 | 2020-10-27 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
6UKT
 
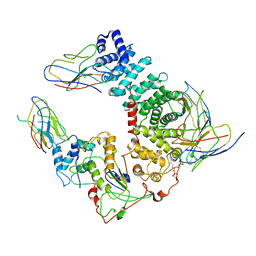 | | Cryo-EM structure of mammalian Ric-8A:Galpha(i):nanobody complex | | 分子名称: | Guanine nucleotide-binding protein G(i) subunit alpha-1, NB8109, NB8117, ... | | 著者 | Mou, T.C, Zhang, K, Johnston, J.D, Chiu, W, Sprang, S.R. | | 登録日 | 2019-10-05 | | 公開日 | 2020-03-11 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | Structure of the G protein chaperone and guanine nucleotide exchange factor Ric-8A bound to G alpha i1.
Nat Commun, 11, 2020
|
|
2I22
 
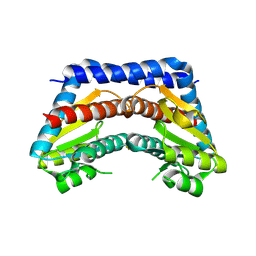 | | Crystal structure of Escherichia coli phosphoheptose isomerase in complex with reaction substrate sedoheptulose 7-phosphate | | 分子名称: | D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, Phosphoheptose isomerase | | 著者 | Blakely, K, Zhang, K, DeLeon, G, Wright, G, Junop, M. | | 登録日 | 2006-08-15 | | 公開日 | 2007-08-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants
J.Biol.Chem., 283, 2008
|
|
6V5C
 
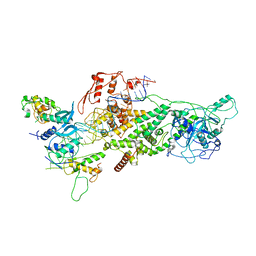 | | Human Drosha and DGCR8 in complex with Primary MicroRNA (MP/RNA complex) - partially docked state | | 分子名称: | Microprocessor complex subunit DGCR8, Pri-miR-16-2 (66-MER), Ribonuclease 3 | | 著者 | Partin, A, Zhang, K, Jeong, B, Herrell, E, Li, S, Chiu, W, Nam, Y. | | 登録日 | 2019-12-04 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA.
Mol.Cell, 78, 2020
|
|
