1BYH
 
 | | MOLECULAR AND ACTIVE-SITE STRUCTURE OF A BACILLUS (1-3,1-4)-BETA-GLUCANASE | | 分子名称: | CALCIUM ION, HYBRID, N-BUTANE, ... | | 著者 | Keitel, T, Heinemann, U. | | 登録日 | 1992-12-31 | | 公開日 | 1993-10-31 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Molecular and active-site structure of a Bacillus 1,3-1,4-beta-glucanase.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
2VTG
 
 | | Crystal Structure of Human Iba2, trigonal crystal form | | 分子名称: | ACETATE ION, IONIZED CALCIUM-BINDING ADAPTER MOLECULE 2, ZINC ION | | 著者 | Schulze, J.O, Quedenau, C, Roske, Y, Turnbull, A, Mueller, U, Heinemann, U, Buessow, K. | | 登録日 | 2008-05-15 | | 公開日 | 2009-07-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural and Functional Characterization of Human Iba Proteins.
FEBS J., 275, 2008
|
|
1GZU
 
 | | Crystal Structure of Human Nicotinamide Mononucleotide Adenylyltransferase in Complex with NMN | | 分子名称: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, NICOTINAMIDE MONONUCLEOTIDE ADENYLYL TRANSFERASE | | 著者 | Werner, E, Ziegler, M, Lerner, F, Schweiger, M, Heinemann, U. | | 登録日 | 2002-06-06 | | 公開日 | 2002-07-05 | | 最終更新日 | 2018-06-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of human nicotinamide mononucleotide adenylyltransferase in complex with NMN.
FEBS Lett., 516, 2002
|
|
1GPP
 
 | |
2XKW
 
 | | LIGAND BINDING DOMAIN OF HUMAN PPAR-GAMMA IN COMPLEX WITH THE AGONIST PIOGLITAZONE | | 分子名称: | (5R)-5-{4-[2-(5-ethylpyridin-2-yl)ethoxy]benzyl}-1,3-thiazolidine-2,4-dione, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Mueller, J.J, Schupp, M, Unger, T, Kintscher, U, Heinemann, U. | | 登録日 | 2010-07-14 | | 公開日 | 2011-08-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Binding Diversity of Pioglitazone by Peroxisome Proliferator-Activated Receptor-Gamma
To be Published
|
|
5OF2
 
 | | The structural versatility of TasA in B. subtilis biofilm formation | | 分子名称: | 1,2-ETHANEDIOL, SULFATE ION, Spore coat-associated protein N | | 著者 | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | 登録日 | 2017-07-10 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5OF1
 
 | | The structural versatility of TasA in B. subtilis biofilm formation | | 分子名称: | 1,2-ETHANEDIOL, 2-HYDROXYBENZOIC ACID, Spore coat-associated protein N | | 著者 | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | 登録日 | 2017-07-10 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5NX0
 
 | | Structure of Spin-labelled T4 lysozyme mutant L115C-R119C-R1 at room temperature | | 分子名称: | Endolysin | | 著者 | Gohlke, U, Loll, B, Consentius, P, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | 登録日 | 2017-05-09 | | 公開日 | 2017-07-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Combining EPR spectroscopy and X-ray crystallography to elucidate the structure and dynamics of conformationally constrained spin labels in T4 lysozyme single crystals.
Phys Chem Chem Phys, 19, 2017
|
|
3ZM0
 
 | | Catalytic domain of human SHP2 | | 分子名称: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | 著者 | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | 登録日 | 2013-02-04 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
3ZM2
 
 | | Catalytic domain of human SHP2 | | 分子名称: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | 著者 | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | 登録日 | 2013-02-04 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
3ZM3
 
 | | Catalytic domain of human SHP2 | | 分子名称: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | 著者 | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | 登録日 | 2013-02-04 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
3ZM1
 
 | | Catalytic domain of human SHP2 | | 分子名称: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | 著者 | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | 登録日 | 2013-02-04 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
4A4I
 
 | | Crystal structure of the human Lin28b cold shock domain | | 分子名称: | GLYCEROL, PROTEIN LIN-28 HOMOLOG B, SULFATE ION | | 著者 | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | 登録日 | 2011-10-14 | | 公開日 | 2012-08-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The Lin28 Cold-Shock Domain Remodels Pre-Let-7 Microrna.
Nucleic Acids Res., 40, 2012
|
|
4AH2
 
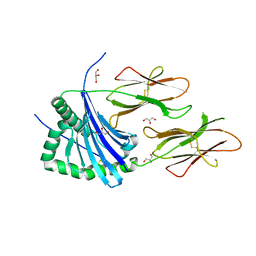 | | HLA-DR1 with covalently linked CLIP106-120 in canonical orientation | | 分子名称: | GLYCEROL, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN GAMMA CHAIN, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN,DRB1-1 BETA CHAIN, ... | | 著者 | Schlundt, A, Guenther, S, Sticht, J, Wieczorek, M, Roske, Y, Heinemann, U, Freund, C. | | 登録日 | 2012-02-03 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Peptide Linkage to the Alpha-Subunit of Mhcii Creates a Stably Inverted Antigen Presentation Complex.
J.Mol.Biol., 423, 2012
|
|
4A75
 
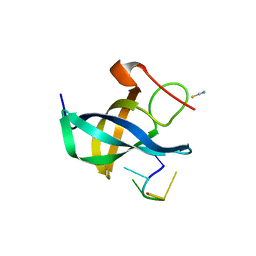 | | The Lin28b Cold shock domain in complex with hexathymidine. | | 分子名称: | 5'-D(*TP*TP*TP*TP*TP*TP)-3', LIN28 COLD SHOCK DOMAIN, THIOCYANATE ION | | 著者 | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | 登録日 | 2011-11-11 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The Lin28 Cold-Shock Domain Remodels Pre-Let-7 Microrna.
Nucleic Acids Res., 40, 2012
|
|
4ZP3
 
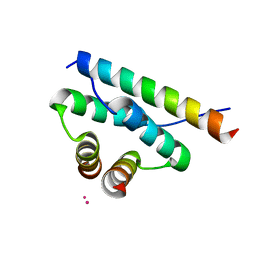 | | AKAP18:PKA-RIIalpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA | | 分子名称: | A-kinase anchor protein 7 isoforms alpha and beta, CADMIUM ION, cAMP-dependent protein kinase type II-alpha regulatory subunit | | 著者 | Goetz, F, Roske, Y, Faelber, K, Zuehlke, K, Autenrieth, K, Kreuchwig, A, Krause, G, Herberg, F.W, Daumke, O, Heinemann, U, Klussmann, E. | | 登録日 | 2015-05-07 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | AKAP18:PKA-RII alpha structure reveals crucial anchor points for recognition of regulatory subunits of PKA.
Biochem.J., 473, 2016
|
|
4ALP
 
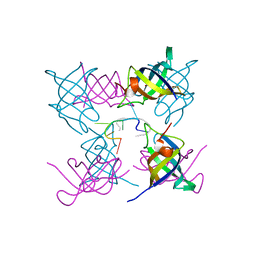 | | The Lin28b Cold shock domain in complex with hexauridine | | 分子名称: | GLYCEROL, HEXA URIDINE, LIN28 ISOFORM B | | 著者 | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | 登録日 | 2012-03-05 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | The Lin28 Cold-Shock Domain Remodels Pre-Let-7 Microrna.
Nucleic Acids Res., 40, 2012
|
|
4AEN
 
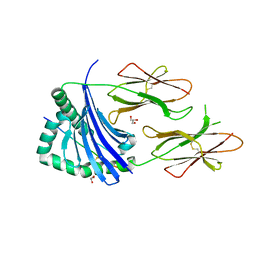 | | HLA-DR1 with covalently linked CLIP106-120 in reversed orientation | | 分子名称: | GLYCEROL, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN GAMMA CHAIN, HLA CLASS II HISTOCOMPATIBILITY ANTIGEN, ... | | 著者 | Schlundt, A, Guenther, S, Sticht, J, Wieczorek, M, Roske, Y, Heinemann, U, Freund, C. | | 登録日 | 2012-01-11 | | 公開日 | 2012-08-01 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Peptide Linkage to the Alpha-Subunit of Mhcii Creates a Stably Inverted Antigen Presentation Complex.
J.Mol.Biol., 423, 2012
|
|
4A76
 
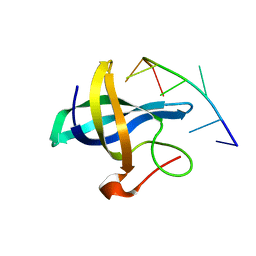 | | The Lin28b Cold shock domain in complex with heptathymidine | | 分子名称: | 5'-D(*TP*TP*TP*TP*TP*TP*TP)-3', LIN28 COLD SHOCK DOMAIN | | 著者 | Mayr, F, Schuetz, A, Doege, N, Heinemann, U. | | 登録日 | 2011-11-11 | | 公開日 | 2012-09-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | The Lin28 Cold-Shock Domain Remodels Pre-Let-7 Microrna.
Nucleic Acids Res., 40, 2012
|
|
6ETE
 
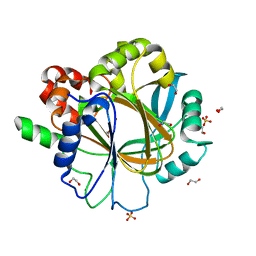 | | Crystal structure of KDM4D with tetrazolhydrazide compound 5 | | 分子名称: | 1,2-ETHANEDIOL, Lysine-specific demethylase 4D, NICKEL (II) ION, ... | | 著者 | Malecki, P.H, Weiss, M.S, Heinemann, U, Link, A. | | 登録日 | 2017-10-26 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.468 Å) | | 主引用文献 | Crystal structure of KDM4D with tetrazolylhydrazide ligand NR128
To be published
|
|
6ETS
 
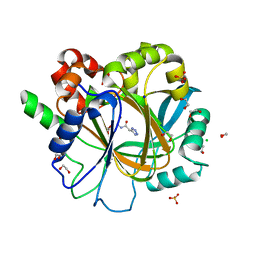 | | Crystal structure of KDM4D with tetrazolhydrazide compound 1 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | 著者 | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | 登録日 | 2017-10-27 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.333 Å) | | 主引用文献 | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6F5R
 
 | | Crystal Structure of KDM4D with GF028 ligand | | 分子名称: | 1,2-ETHANEDIOL, 2-(3-oxidanylpropylamino)pyridine-4-carboxylic acid, CHLORIDE ION, ... | | 著者 | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | 登録日 | 2017-12-02 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.607 Å) | | 主引用文献 | Crystal Structure of KDM4D with GF028 ligand
To be published
|
|
6F5S
 
 | | Crystal Structure of KDM4D with tetrazole ligand GF049 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | 著者 | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | 登録日 | 2017-12-02 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Crystal Structure of KDM4D with tetrazole ligand GF049
To be published
|
|
6F7K
 
 | | Crystal structure of Dettilon tailspike protein (gp208) | | 分子名称: | 1,2-ETHANEDIOL, Tailspike, alpha-D-galactopyranose-(1-6)-beta-D-mannopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-alpha-D-galactopyranose, ... | | 著者 | Roske, Y, Heinemann, U. | | 登録日 | 2017-12-11 | | 公開日 | 2018-12-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Time-resolved DNA release from an O-antigen-specificSalmonellabacteriophage with a contractile tail.
J.Biol.Chem., 294, 2019
|
|
6ETT
 
 | | Crystal structure of KDM4D with tetrazole compound 4 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | 著者 | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | 登録日 | 2017-10-27 | | 公開日 | 2019-02-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.257 Å) | | 主引用文献 | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
