8VH1
 
 | | CH235.12 Fab bound to the HIV-1 CH505.M5 SOSIP | | 分子名称: | CH235.I60 Fab Heavy Chain, CH235.I60 Fab Light Chain, CH505.M5 SOSIP gp120, ... | | 著者 | Henderson, R, Acharya, P. | | 登録日 | 2023-12-30 | | 公開日 | 2024-10-02 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (4.08 Å) | | 主引用文献 | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies.
Nat Commun, 15, 2024
|
|
7F3P
 
 | | Crystal structure of a nadp-dependent alcohol dehydrogenase mutant in apo form | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent isopropanol dehydrogenase, ZINC ION | | 著者 | Han, X, Bi, Y, Wei, H.L, Gao, J, Li, Q, Qu, G, Sun, Z.T, Liu, W.D. | | 登録日 | 2021-06-16 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Unlocking the Stereoselectivity and Substrate Acceptance of Enzymes: Proline-Induced Loop Engineering Test.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
4N6E
 
 | | Crystal structure of Amycolatopsis orientalis BexX/CysO complex | | 分子名称: | Putative thiosugar synthase, SULFATE ION, ThiS/MoaD family protein | | 著者 | Zhang, X, Zhang, Y, Kinsland, C, Sasaki, E, Sun, H.G, Lu, M.J, Liu, T, Ou, A, Li, J, Chen, Y, Liu, H, Ealick, S.E. | | 登録日 | 2013-10-11 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Co-opting sulphur-carrier proteins from primary metabolic pathways for 2-thiosugar biosynthesis.
Nature, 509, 2014
|
|
4DA1
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase phosphatase with Mg (II) ions at the active site | | 分子名称: | BETA-MERCAPTOETHANOL, MAGNESIUM ION, Protein phosphatase 1K, ... | | 著者 | Brautigam, C.A, Chuang, J.L, Chuang, D.T. | | 登録日 | 2012-01-12 | | 公開日 | 2012-02-08 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.383 Å) | | 主引用文献 | Structural and biochemical characterization of human mitochondrial branched-chain alpha-ketoacid dehydrogenase phosphatase.
J.Biol.Chem., 287, 2012
|
|
3R2C
 
 | |
7K93
 
 | |
5BT1
 
 | | histone chaperone Hif1 playing with histone H2A-H2B dimer | | 分子名称: | HAT1-interacting factor 1, Histone H2A.1, Histone H2B.1 | | 著者 | Liu, H, Zhang, M, Gao, Y, Teng, M, Niu, L. | | 登録日 | 2015-06-02 | | 公開日 | 2016-10-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Structural Insights into the Association of Hif1 with Histones H2A-H2B Dimer and H3-H4 Tetramer
Structure, 24, 2016
|
|
9AVO
 
 | |
9AWE
 
 | |
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | 分子名称: | Methyltransferase | | 著者 | Chen, H, Lin, S, Lu, G.W. | | 登録日 | 2022-09-21 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
8GY9
 
 | |
5CFF
 
 | |
8WRJ
 
 | | glycosyltransferase UGT74AN3 | | 分子名称: | 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | 著者 | Huang, W, Long, F. | | 登録日 | 2023-10-15 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8WRK
 
 | | glycosyltransferase UGT74AN3 | | 分子名称: | Glycosyltransferase, PICEATANNOL, URIDINE-5'-DIPHOSPHATE | | 著者 | Huang, W, Long, F. | | 登録日 | 2023-10-15 | | 公開日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8I6Q
 
 | |
8I5X
 
 | | Structure of human Nav1.7 in complex with Vinpocetine | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Wu, Q.R, Yan, N. | | 登録日 | 2023-01-26 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5B
 
 | | Structure of human Nav1.7 in complex with bupivacaine | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Wu, Q.R, Yan, N. | | 登録日 | 2023-01-24 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5Y
 
 | | Structure of human Nav1.7 in complex with vixotrigine | | 分子名称: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Wu, Q.R, Yan, N. | | 登録日 | 2023-01-26 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
7WKA
 
 | |
8I5G
 
 | | Structure of human Nav1.7 in complex with PF-05089771 | | 分子名称: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | 著者 | Wu, Q.R, Yan, N. | | 登録日 | 2023-01-25 | | 公開日 | 2023-06-14 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
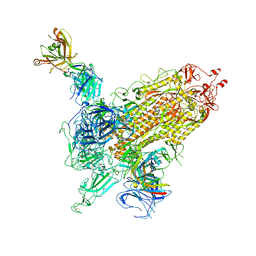
7WK9
 
 | |
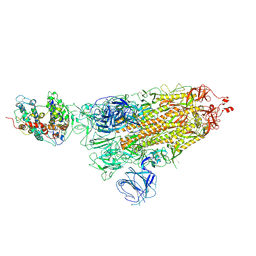
7WK4
 
 | |
7WK5
 
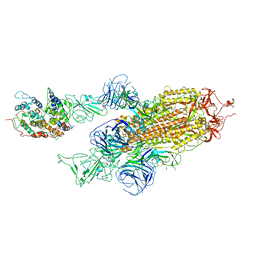 | | Cryo-EM structure of Omicron S-ACE2, C2 state | | 分子名称: | Angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Han, W.Y, Wang, Y.F. | | 登録日 | 2022-01-08 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Molecular basis of SARS-CoV-2 Omicron variant receptor engagement and antibody evasion and neutralization
Biorxiv, 2022
|
|
7WK8
 
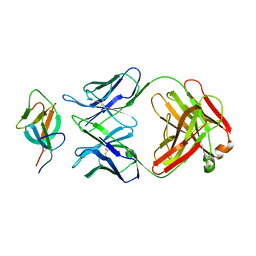 | |
7W0G
 
 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist H11 | | 分子名称: | 2-[2,6-dimethyl-4-[[5-oxidanylidene-4-[4-(trifluoromethyloxy)phenyl]-1,2,4-triazol-1-yl]methyl]phenoxy]-2-methyl-propanoic acid, Peroxisome proliferator-activated receptor delta | | 著者 | Dai, L, Sun, H.B, Yuan, H.L, Feng, Z.Q. | | 登録日 | 2021-11-18 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.443 Å) | | 主引用文献 | Design, Synthesis, and Biological Evaluation of Triazolone Derivatives as Potent PPAR alpha / delta Dual Agonists for the Treatment of Nonalcoholic Steatohepatitis.
J.Med.Chem., 65, 2022
|
|
