7XSS
 
 | | Structure of Craspase-CTR | | 分子名称: | CHAT domain protein, RAMP superfamily protein, RNA (34-MER), ... | | 著者 | Feng, Y, Zang, L.X. | | 登録日 | 2022-05-15 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Target RNA activates the protease activity of Craspase to confer antiviral defense.
Mol.Cell, 82, 2022
|
|
7E8M
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with mutated RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | 著者 | Wang, X.Q, Zhang, L.Q, Ge, J.W, Wang, R.K, Lan, J. | | 登録日 | 2021-03-02 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species.
Immunity, 54, 2021
|
|
7W5S
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (II) ION, ... | | 著者 | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | 登録日 | 2021-11-30 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5T
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | 分子名称: | 2-OXOGLUTARIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | 登録日 | 2021-11-30 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
7W5V
 
 | | A nonheme iron- and alpha-ketoglutarate- dependent halogenase that catalyzes nucleotide substrates | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | 著者 | Dai, L.H, Zhang, X, Hu, Y.M, Huang, J.W, Chen, C.C, Guo, R.T. | | 登録日 | 2021-11-30 | | 公開日 | 2022-04-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural and Functional Insights into a Nonheme Iron- and alpha-Ketoglutarate-Dependent Halogenase That Catalyzes Chlorination of Nucleotide Substrates.
Appl.Environ.Microbiol., 88, 2022
|
|
1AQK
 
 | |
7B14
 
 | |
5ZMA
 
 | |
7B18
 
 | | SARS-CoV-spike bound to two neutralising nanobodies | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 VHH E, ... | | 著者 | Hallberg, B.M, Das, H. | | 登録日 | 2020-11-24 | | 公開日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
6KA2
 
 | | Crystal structure of a Thebaine synthase from Papaver somniferum in complex with TBN | | 分子名称: | (4R,7aR,12bS)-7,9-dimethoxy-3-methyl-2,4,7a,13-tetrahydro-1H-4,12-methanobenzofuro[3,2-e]isoquinoline, Thebaine synthase 2 | | 著者 | Xue, J, Yu, X.J, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2019-06-20 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural insights into thebaine synthase 2 catalysis.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
7WCY
 
 | | Crystal Structure of H-2Kb with Cryptosporidium parvum gp40/15 epitope | | 分子名称: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | 著者 | Wang, Y.L, Gao, M.H, Zhang, L.X, Fan, S.H. | | 登録日 | 2021-12-20 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structural Analyses of a Dominant Cryptosporidium parvum Epitope Presented by H-2K b Offer New Options To Combat Cryptosporidiosis.
Mbio, 14, 2023
|
|
2VD4
 
 | | Structure of small-molecule inhibitor of Glmu from Haemophilus influenzae reveals an allosteric binding site | | 分子名称: | 4-chloro-N-(3-methoxypropyl)-N-[(3S)-1-(2-phenylethyl)piperidin-3-yl]benzamide, BIFUNCTIONAL PROTEIN GLMU, MAGNESIUM ION, ... | | 著者 | Mochalkin, I, Lightle, S, McDowell, L. | | 登録日 | 2007-09-28 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of a Small-Molecule Inhibitor Complexed with Glmu from Haemophilus Influenzae Reveals an Allosteric Binding Site.
Protein Sci., 17, 2008
|
|
1SYS
 
 | | Crystal structure of HLA, B*4403, and peptide EEPTVIKKY | | 分子名称: | Beta-2-microglobulin, Sorting nexin 5, leukocyte antigen (HLA) class I molecule | | 著者 | Zernich, D, Purcell, A.W, Macdonald, W.A, Kjer-Nielsen, L, Ely, L.K, Laham, N, Crockford, T, Mifsud, N.A, Tait, B.D, Holdsworth, R, Brooks, A.G, Bottomley, S.P, Beddoe, T, Peh, C.A, Rossjohn, J, McCluskey, J. | | 登録日 | 2004-04-01 | | 公開日 | 2004-10-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion
J.Exp.Med., 200, 2004
|
|
7Y9G
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 in complex with pyrophosphate | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2022-06-24 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7Y9H
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diterpene synthase VenA, ... | | 著者 | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | 登録日 | 2022-06-24 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
6KA3
 
 | | Crystal structure of a Thebaine synthase from Papaver somniferum | | 分子名称: | PALMITIC ACID, SULFATE ION, Thebaine synthase 2 | | 著者 | Xue, J, Yu, X.J, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | 登録日 | 2019-06-20 | | 公開日 | 2020-06-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.951 Å) | | 主引用文献 | Structural insights into thebaine synthase 2 catalysis.
Biochem.Biophys.Res.Commun., 529, 2020
|
|
3RDH
 
 | | X-ray induced covalent inhibition of 14-3-3 | | 分子名称: | 14-3-3 protein zeta/delta, 4-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]benzoic acid, NICKEL (II) ION | | 著者 | Horton, J.R, Upadhyay, A.K, Fu, H, Cheng, X. | | 登録日 | 2011-04-01 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Discovery and structural characterization of a small molecule 14-3-3 protein-protein interaction inhibitor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7VJQ
 
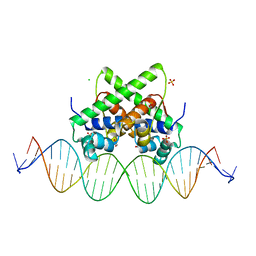 | | Pectobacterium phage ZF40 apo-aca2 complexed with 26bp DNA substrate | | 分子名称: | CHLORIDE ION, DNA (27-MER), GLYCEROL, ... | | 著者 | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | 登録日 | 2021-09-28 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7VJO
 
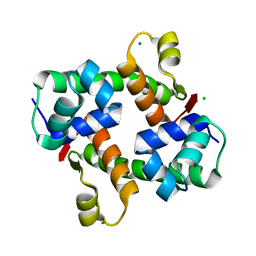 | | Pectobacterium phage ZF40 apo-Aca2 | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, anti-CRISPR-associated protein Aca2 | | 著者 | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | 登録日 | 2021-09-28 | | 公開日 | 2021-10-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7VJN
 
 | |
7VJM
 
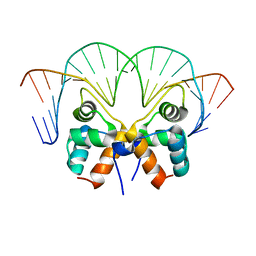 | | Aca1 in complex with 19bp palindromic DNA substrate | | 分子名称: | DNA (5'-D(*AP*TP*TP*AP*GP*GP*CP*AP*CP*AP*TP*TP*GP*TP*GP*CP*CP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*GP*GP*CP*AP*CP*AP*AP*TP*GP*TP*GP*CP*CP*TP*AP*A)-3'), anti-CRISPR-associated protein Aca1 | | 著者 | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | 登録日 | 2021-09-28 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7VJP
 
 | | Selenomethionine-derived Pectobacterium phage ZF40 apo-Aca2 | | 分子名称: | SULFATE ION, anti-CRISPR-associated protein Aca2 | | 著者 | Liu, Y.H, Zhang, L.S, Wu, B.X, Huang, H.D. | | 登録日 | 2021-09-28 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-02-02 | | 実験手法 | X-RAY DIFFRACTION (1.594 Å) | | 主引用文献 | Structural basis for anti-CRISPR repression mediated by bacterial operon proteins Aca1 and Aca2.
J.Biol.Chem., 297, 2021
|
|
7VH8
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332 | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Zhao, Y, Zhang, Q, Yang, H, Rao, Z. | | 登録日 | 2021-09-21 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332.
Protein Cell, 13, 2022
|
|
2W0W
 
 | | Crystal structure of Glmu from Haemophilus influenzae in complex with quinazoline inhibitor 2 | | 分子名称: | GLUCOSAMINE-1-PHOSPHATE N-ACETYLTRANSFERASE, N-{6-(CYCLOPROPYLMETHOXY)-7-METHOXY-2-[6-(2-METHYLPROPYL)-5-OXO-3,4,5,6-TETRAHYDRO-2,6-NAPHTHYRIDIN-2(1H)-YL]QUINAZOLIN-4-YL}-2,2,2-TRIFLUOROETHANESULFONAMIDE, TETRAETHYLENE GLYCOL | | 著者 | Mochalkin, I, Melnick, M. | | 登録日 | 2008-10-10 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Discovery and Initial Sar of Quinazoline Inhibitors of Glmu from Haemophilus Influenzae
To be Published
|
|
3SQJ
 
 | |
