2FBT
 
 | | WRN exonuclease | | 分子名称: | ACETIC ACID, Werner syndrome helicase | | 著者 | Perry, J.J. | | 登録日 | 2005-12-10 | | 公開日 | 2006-04-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2FBV
 
 | | WRN exonuclease, Mn complex | | 分子名称: | MANGANESE (II) ION, Werner syndrome helicase | | 著者 | Perry, J.J. | | 登録日 | 2005-12-10 | | 公開日 | 2006-04-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing.
Nat.Struct.Mol.Biol., 13, 2006
|
|
7CPV
 
 | | Cryo-EM structure of 80S ribosome from mouse testis | | 分子名称: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | 著者 | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | 登録日 | 2020-08-08 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
7CPU
 
 | | Cryo-EM structure of 80S ribosome from mouse kidney | | 分子名称: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | 著者 | Huo, Y.G, He, X, Jiang, T, Qin, Y, Guo, X.J, Sha, J.H. | | 登録日 | 2020-08-08 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.82 Å) | | 主引用文献 | A male germ-cell-specific ribosome controls male fertility.
Nature, 612, 2022
|
|
6BY9
 
 | | Crystal structure of EHMT1 | | 分子名称: | Histone-lysine N-methyltransferase EHMT1, UNKNOWN ATOM OR ION | | 著者 | Dong, A, Wei, Y, Li, A, Tempel, W, Han, S, Sunnerhagen, M, Penn, L, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | 登録日 | 2017-12-20 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of EHMT1
to be published
|
|
4ICR
 
 | | Structural basis for substrate recognition and reaction mechanism of bacterial aminopeptidase peps | | 分子名称: | Aminopeptidase PepS, CACODYLATE ION, ZINC ION | | 著者 | Lee, S, Kim, K.K, Ta, M.H. | | 登録日 | 2012-12-11 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structure-based elucidation of the regulatory mechanism for aminopeptidase activity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4ICQ
 
 | |
4ICS
 
 | | Crystal structure of PepS from Streptococcus pneumoniae in complex with a substrate | | 分子名称: | Aminopeptidase PepS, GLYCINE, TRYPTOPHAN, ... | | 著者 | Lee, S, Kim, K.K, Ta, M.H. | | 登録日 | 2012-12-11 | | 公開日 | 2013-10-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structure-based elucidation of the regulatory mechanism for aminopeptidase activity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2V89
 
 | |
1F46
 
 | | THE BACTERIAL CELL-DIVISION PROTEIN ZIPA AND ITS INTERACTION WITH AN FTSZ FRAGMENT REVEALED BY X-RAY CRYSTALLOGRAPHY | | 分子名称: | CELL DIVISION PROTEIN ZIPA | | 著者 | Mosyak, L, Zhang, Y, Glasfeld, E, Stahl, M, Somers, W.S. | | 登録日 | 2000-06-07 | | 公開日 | 2001-06-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography.
EMBO J., 19, 2000
|
|
1F47
 
 | | THE BACTERIAL CELL-DIVISION PROTEIN ZIPA AND ITS INTERACTION WITH AN FTSZ FRAGMENT REVEALED BY X-RAY CRYSTALLOGRAPHY | | 分子名称: | CELL DIVISION PROTEIN FTSZ, CELL DIVISION PROTEIN ZIPA | | 著者 | Mosyak, L, Zhang, Y, Glasfeld, E, Stahl, M, Somers, W.S. | | 登録日 | 2000-06-07 | | 公開日 | 2001-06-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography.
EMBO J., 19, 2000
|
|
6ISS
 
 | | Lignin peroxidase H8 triple mutant S49C/A67C/H239 | | 分子名称: | CALCIUM ION, Ligninase H8, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Seo, H, Son, H, Kim, K.-J. | | 登録日 | 2018-11-19 | | 公開日 | 2019-11-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Extra disulfide and ionic salt bridge improves the thermostability of lignin peroxidase H8 under acidic condition
Enzyme.Microb.Technol., 148, 2021
|
|
1TBJ
 
 | | H141A mutant of rat liver arginase I | | 分子名称: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | 著者 | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | 登録日 | 2004-05-20 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
7WU5
 
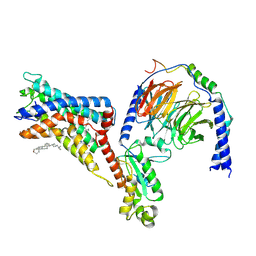 | | Cryo-EM structure of the adhesion GPCR ADGRF1(H565A/T567A) in complex with miniGi | | 分子名称: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2022-02-05 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU3
 
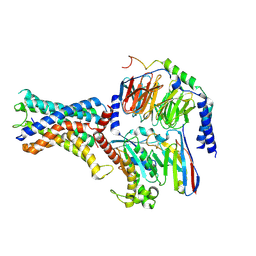 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGs | | 分子名称: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2022-02-05 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU4
 
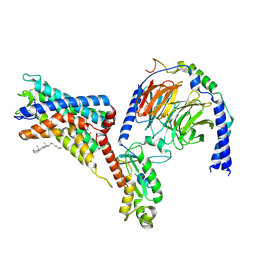 | | Cryo-EM structure of the adhesion GPCR ADGRF1 in complex with miniGi | | 分子名称: | Adhesion G-protein coupled receptor F1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2022-02-05 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
7WU2
 
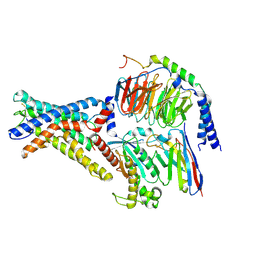 | | Cryo-EM structure of the adhesion GPCR ADGRD1 in complex with miniGs | | 分子名称: | Adhesion G-protein coupled receptor D1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Qu, X, Qiu, N, Wang, M, Zhao, Q, Wu, B. | | 登録日 | 2022-02-05 | | 公開日 | 2022-04-27 | | 最終更新日 | 2022-05-11 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis of tethered agonism of the adhesion GPCRs ADGRD1 and ADGRF1.
Nature, 604, 2022
|
|
1TA1
 
 | | H141C mutant of rat liver arginase I | | 分子名称: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | 著者 | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | 登録日 | 2004-05-19 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
1TBH
 
 | | H141D mutant of rat liver arginase I | | 分子名称: | Arginase 1, MANGANESE (II) ION | | 著者 | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | 登録日 | 2004-05-20 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
1TBL
 
 | | H141N mutant of rat liver arginase I | | 分子名称: | Arginase 1, MANGANESE (II) ION | | 著者 | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | 登録日 | 2004-05-20 | | 公開日 | 2005-08-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
7EEJ
 
 | | Complex structure of glycoside hydrolase family 12 beta-1,3-1,4-glucanase with cellobiose | | 分子名称: | GLYCEROL, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | 著者 | Jiang, Z.Q, Ma, J.W. | | 登録日 | 2021-03-18 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.47798049 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7EE2
 
 | | Structural insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.CQ31 | | 分子名称: | GLYCEROL, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | 著者 | Jiang, Z.Q, Ma, J. | | 登録日 | 2021-03-17 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.37011635 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7EEE
 
 | | Complex structure of glycoside hydrolase family 12 beta-1,3-1,4-glucanase with gentiobiose | | 分子名称: | GLYCEROL, beta-D-mannopyranose-(1-6)-beta-D-mannopyranose, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | 著者 | Jiang, Z.Q, Ma, J.W. | | 登録日 | 2021-03-18 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.660792 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7BWJ
 
 | | crystal structure of SARS-CoV-2 antibody with RBD | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | 著者 | Wang, X, Ge, J. | | 登録日 | 2020-04-14 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Human neutralizing antibodies elicited by SARS-CoV-2 infection.
Nature, 584, 2020
|
|
8DD5
 
 | | Crystal structure of KAT6A in complex with inhibitor CTx-648 (PF-9363) | | 分子名称: | 2,6-dimethoxy-N-{4-methoxy-6-[(1H-pyrazol-1-yl)methyl]-1,2-benzoxazol-3-yl}benzene-1-sulfonamide, Histone acetyltransferase KAT6A, ZINC ION | | 著者 | Greasley, S.E, Johnson, E, Brodsky, O. | | 登録日 | 2022-06-17 | | 公開日 | 2023-07-05 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Targeting KAT6A/KAT6B dependencies in breast cancer with a novel selective, orally bioavailable KAT6 inhibitor, CTx-648/PF-9363
To Be Published
|
|
