7V4N
 
 | |
7V4P
 
 | |
7V4F
 
 | |
6K5O
 
 | | Development of Novel Lithocholic Acid Derivatives as Vitamin D Receptor Agonists | | 分子名称: | (4~{R})-4-[(3~{R},5~{R},8~{R},9~{S},10~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3-methylsulfonyloxy-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | 著者 | Masuno, H, Kagechika, H, Ito, N. | | 登録日 | 2019-05-29 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Development of novel lithocholic acid derivatives as vitamin D receptor agonists.
Bioorg.Med.Chem., 27, 2019
|
|
2K86
 
 | | Solution Structure of FOXO3a Forkhead domain | | 分子名称: | Forkhead box protein O3 | | 著者 | Wang, F, Marshall, C.B, Li, G, Plevin, M.J, Ikura, M. | | 登録日 | 2008-09-02 | | 公開日 | 2008-10-14 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Biochemical and structural characterization of an intramolecular interaction in FOXO3a and its binding with p53.
J.Mol.Biol., 384, 2008
|
|
5AVE
 
 | | The ligand binding domain of Mlp37 with serine | | 分子名称: | Methyl-accepting chemotaxis (MCP) signaling domain protein, SERINE | | 著者 | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | 登録日 | 2015-06-15 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
5B4H
 
 | | Crystal structure of I86D mutant of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin (data 1) | | 分子名称: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | 著者 | Hagiwara, Y, Wada, K, Irikawa, T, Unno, M, Fukuyama, K, Sugishima, M. | | 登録日 | 2016-04-04 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.11 Å) | | 主引用文献 | Atomic-resolution structure of the phycocyanobilin:ferredoxin oxidoreductase I86D mutant in complex with fully protonated biliverdin
FEBS Lett., 590, 2016
|
|
5B4J
 
 | | Crystal structure of I86D mutant of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin (data 3) | | 分子名称: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | 著者 | Hagiwara, Y, Wada, K, Irikawa, T, Unno, M, Fukuyama, K, Sugishima, M. | | 登録日 | 2016-04-04 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Atomic-resolution structure of the phycocyanobilin:ferredoxin oxidoreductase I86D mutant in complex with fully protonated biliverdin
FEBS Lett., 590, 2016
|
|
5AVF
 
 | | The ligand binding domain of Mlp37 with taurine | | 分子名称: | 2-AMINOETHANESULFONIC ACID, Methyl-accepting chemotaxis (MCP) signaling domain protein | | 著者 | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | 登録日 | 2015-06-15 | | 公開日 | 2016-06-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
5B4I
 
 | | Crystal structure of I86D mutant of phycocyanobilin:ferredoxin oxidoreductase in complex with biliverdin (data 2) | | 分子名称: | BILIVERDINE IX ALPHA, Phycocyanobilin:ferredoxin oxidoreductase | | 著者 | Hagiwara, Y, Wada, K, Irikawa, T, Unno, M, Fukuyama, K, Sugishima, M. | | 登録日 | 2016-04-04 | | 公開日 | 2017-03-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.11 Å) | | 主引用文献 | Atomic-resolution structure of the phycocyanobilin:ferredoxin oxidoreductase I86D mutant in complex with fully protonated biliverdin
FEBS Lett., 590, 2016
|
|
2ZXM
 
 | | A New Class of Vitamin D Receptor Ligands that Induce Structural Rearrangement of the Ligand-binding Pocket | | 分子名称: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,3S)-3-(2-hydroxyethyl)heptan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | 著者 | Nakabayashi, M, Ikura, T, Ito, N. | | 登録日 | 2009-01-04 | | 公開日 | 2009-02-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | A New Class of Vitamin D Analogues that Induce Structural Rearrangement of the Ligand-Binding Pocket of the Receptor
J.Med.Chem., 52, 2009
|
|
2ZXN
 
 | | A New Class of Vitamin D Receptor Ligands that Induce Structural Rearrangement of the Ligand-binding Pocket | | 分子名称: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2S,3S)-3-(2-hydroxyethyl)heptan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | 著者 | Nakabayashi, M, Ikura, T, Ito, N. | | 登録日 | 2009-01-04 | | 公開日 | 2009-02-17 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A New Class of Vitamin D Analogues that Induce Structural Rearrangement of the Ligand-Binding Pocket of the Receptor
J.Med.Chem., 52, 2009
|
|
1BDM
 
 | |
3WEX
 
 | | Crystal structure of HLA-DP5 in complex with Cry j 1-derived peptide (residues 214-222) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MHC class II antigen | | 著者 | Kusano, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | 登録日 | 2013-07-16 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for the specific recognition of the major antigenic peptide from the Japanese cedar pollen allergen Cry j 1 by HLA-DP5
J. Mol. Biol., 426, 2014
|
|
2D4D
 
 | | The Crystal Structure of human beta2-microglobulin, L39W W60F W95F Mutant | | 分子名称: | Beta-2-microglobulin, SODIUM ION | | 著者 | Iwata, K, Matsuura, T, Nakagawa, A, Goto, Y. | | 登録日 | 2005-10-17 | | 公開日 | 2006-08-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Conformation of Amyloid Fibrils of beta2-Microglobulin Probed by Tryptophan Mutagenesis
J.Biol.Chem., 281, 2006
|
|
7WDT
 
 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with GlcNAc-6S | | 分子名称: | 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose, 2-acetamido-2-deoxy-6-O-sulfo-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ... | | 著者 | Yamada, C, Kashima, T, Fushinobu, S, Katoh, T, Katayama, T. | | 登録日 | 2021-12-22 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
7WDU
 
 | | 6-sulfo-beta-D-N-acetylglucosaminidase from Bifidobacterium bifidum in complex with PUGNAc-6S | | 分子名称: | Beta-N-acetylhexosaminidase, CALCIUM ION, [[(3R,4R,5S,6R)-3-acetamido-4,5-bis(oxidanyl)-6-(sulfooxymethyl)oxan-2-ylidene]amino] N-phenylcarbamate | | 著者 | Kashima, T, Yamada, C, Fushinobu, S, Katoh, T, Katayama, T. | | 登録日 | 2021-12-22 | | 公開日 | 2022-12-28 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem.
Nat.Chem.Biol., 19, 2023
|
|
2D4F
 
 | | The Crystal Structure of human beta2-microglobulin | | 分子名称: | Beta-2-microglobulin, SODIUM ION | | 著者 | Iwata, K, Matsuura, T, Nakagawa, A, Goto, Y. | | 登録日 | 2005-10-18 | | 公開日 | 2006-08-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Conformation of Amyloid Fibrils of beta2-Microglobulin Probed by Tryptophan Mutagenesis
J.Biol.Chem., 281, 2006
|
|
5B1W
 
 | |
5B1X
 
 | |
1Y7T
 
 | | Crystal structure of NAD(H)-depenent malate dehydrogenase complexed with NADPH | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Tomita, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | 登録日 | 2004-12-10 | | 公開日 | 2005-08-02 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of NAD-dependent malate dehydrogenase complexed with NADP(H)
Biochem.Biophys.Res.Commun., 334, 2005
|
|
1BMD
 
 | |
7CD0
 
 | | Crystal structure of the 2-iodoporphobilinogen-bound ES2 intermediate form of human hydroxymethylbilane synthase | | 分子名称: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, 3-[5-(aminomethyl)-4-(carboxymethyl)-2-iodo-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | 著者 | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | 登録日 | 2020-06-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
7CCZ
 
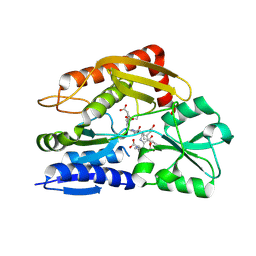 | | Crystal structure of the ES2 intermediate form of human hydroxymethylbilane synthase | | 分子名称: | 3-[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-5-[[4-(2-hydroxy-2-oxoethyl)-3-(3-hydroxy-3-oxopropyl)-5-methyl-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-3-(3-hydroxy-3-oxopropyl)-1~{H}-pyrrol-2-yl]methyl]-1~{H}-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | 著者 | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | 登録日 | 2020-06-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
7CCY
 
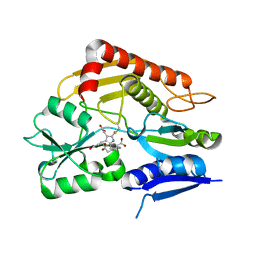 | | Crystal structure of the 2-iodoporphobilinogen-bound holo form of human hydroxymethylbilane synthase | | 分子名称: | 3-[5-(aminomethyl)-4-(carboxymethyl)-2-iodo-1H-pyrrol-3-yl]propanoic acid, 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, Porphobilinogen deaminase | | 著者 | Sato, H, Sugishima, M, Wada, K, Hirabayashi, K, Tsukaguchi, M. | | 登録日 | 2020-06-18 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of hydroxymethylbilane synthase complexed with a substrate analog: a single substrate-binding site for four consecutive condensation steps.
Biochem.J., 478, 2021
|
|
