7XHN
 
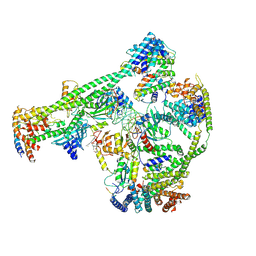 | | Structure of human inner kinetochore CCAN-DNA complex | | 分子名称: | CENP-W, Centromere protein C, Centromere protein H, ... | | 著者 | Sun, L.F, Tian, T, Wang, C.L, Yang, Z.S, Zang, J.Y. | | 登録日 | 2022-04-09 | | 公開日 | 2023-01-25 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Structural insights into human CCAN complex assembled onto DNA.
Cell Discov, 8, 2022
|
|
7D8I
 
 | |
7D8G
 
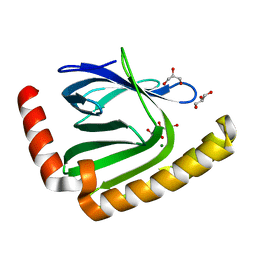 | |
7D8Q
 
 | | The structure of nucleotide phosphatase Sa1684 complex with GDP analogue from Staphylococcus aureus | | 分子名称: | MAGNESIUM ION, UPF0374 protein SAB1800c, [(2R,3R,4S,5S)-5-(2-azanyl-6-oxidanyl-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl bis(oxidanyl)phosphinothioyl hydrogen phosphate | | 著者 | Wang, Z, Li, X. | | 登録日 | 2020-10-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The structural mechanism for the nucleoside tri- and diphosphate hydrolysis activity of Ntdp from Staphylococcus aureus.
Febs J., 288, 2021
|
|
7D8L
 
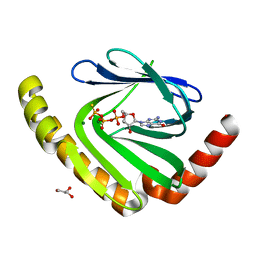 | |
7VGZ
 
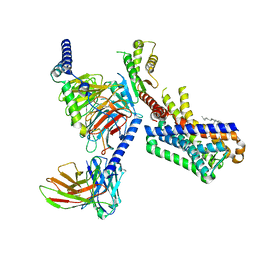 | | MT1-remalteon-Gi complex | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, Q.G, Lu, Q.Y. | | 登録日 | 2021-09-20 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of the ligand binding and signaling mechanism of melatonin receptors.
Nat Commun, 13, 2022
|
|
7VH0
 
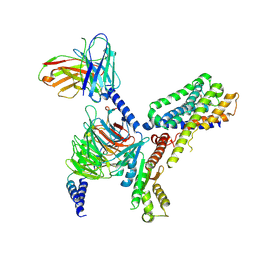 | | MT2-remalteon-Gi complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, Melatonin receptor type 1B, ... | | 著者 | Wang, Q.G, Lu, Q.Y. | | 登録日 | 2021-09-20 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.46 Å) | | 主引用文献 | Structural basis of the ligand binding and signaling mechanism of melatonin receptors.
Nat Commun, 13, 2022
|
|
7VGY
 
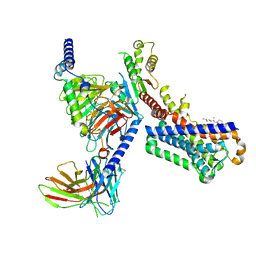 | | Melatonin receptor1-2-Iodomelatonin-Gicomplex | | 分子名称: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, Q.G, Lu, Q.Y. | | 登録日 | 2021-09-20 | | 公開日 | 2022-03-02 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of the ligand binding and signaling mechanism of melatonin receptors.
Nat Commun, 13, 2022
|
|
7YF1
 
 | |
4JWG
 
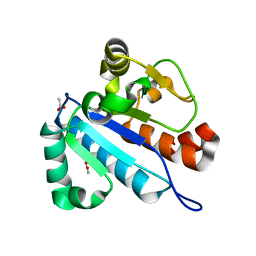 | | Crystal structure of spTrm10(74) | | 分子名称: | ACETIC ACID, tRNA (guanine(9)-N1)-methyltransferase | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2013-03-27 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
4JR7
 
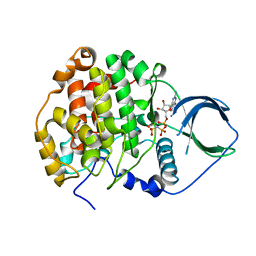 | | Crystal structure of scCK2 alpha in complex with GMPPNP | | 分子名称: | Casein kinase II subunit alpha, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Liu, H. | | 登録日 | 2013-03-21 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LFI
 
 | | Crystal structure of scCK2 alpha in complex with GMPPNP | | 分子名称: | Casein kinase II subunit alpha, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Liu, H. | | 登録日 | 2013-06-27 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4M36
 
 | |
4JQE
 
 | | Crystal structure of scCK2 alpha in complex with AMPPN | | 分子名称: | Casein kinase II subunit alpha, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Liu, H. | | 登録日 | 2013-03-20 | | 公開日 | 2014-03-05 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JWF
 
 | | Crystal structure of spTrm10(74)-SAH complex | | 分子名称: | ACETIC ACID, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine(9)-N1)-methyltransferase | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2013-03-27 | | 公開日 | 2013-10-16 | | 最終更新日 | 2014-02-12 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
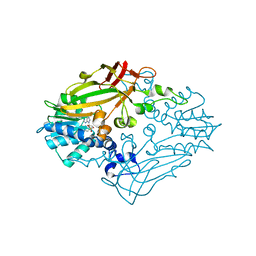
4M37
 
 | |
4JWH
 
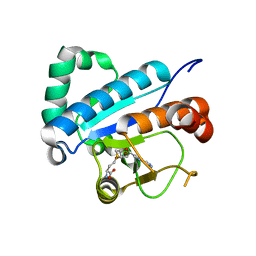 | | Crystal structure of spTrm10(Full length)-SAH complex | | 分子名称: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine(9)-N1)-methyltransferase | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2013-03-27 | | 公開日 | 2013-10-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
4JWJ
 
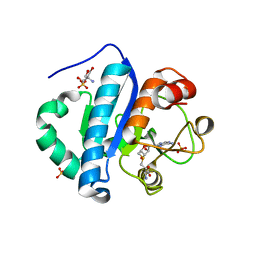 | | Crystal structure of scTrm10(84)-SAH complex | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | 著者 | Yan, W, Shao, Z. | | 登録日 | 2013-03-27 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
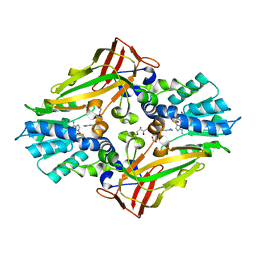
4M38
 
 | |
4MWH
 
 | | Crystal structure of scCK2 alpha in complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Casein kinase II subunit alpha, MAGNESIUM ION, ... | | 著者 | Liu, H. | | 登録日 | 2013-09-24 | | 公開日 | 2013-11-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | The multiple nucleotide-divalent cation binding modes of Saccharomyces cerevisiae CK2 alpha indicate a possible co-substrate hydrolysis product (ADP/GDP) release pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3FE5
 
 | | Crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from bovine kidney | | 分子名称: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION | | 著者 | Dilovic, I, Gliubich, F, Malpeli, G, Zanotti, G, Matkovic-Calogovic, D. | | 登録日 | 2008-11-27 | | 公開日 | 2009-06-09 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Crystal structure of bovine 3-hydroxyanthranilate 3,4-dioxygenase.
Biopolymers, 2009
|
|
303D
 
 | | META-HYDROXY ANALOGUE OF HOECHST 33258 ('HYDROXYL OUT' CONFORMATION) BOUND TO D(CGCGAATTCGCG)2 | | 分子名称: | 3-[5-[5-(4-METHYL-PIPERAZIN-1-YL)-1H-IMIDAZO[4,5-B]PYRIDIN-2-YL]-BENZIMIDAZOL-2-YL]-PHENOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | 著者 | Clark, G.R, Squire, C.J, Gray, E.J, Leupin, W, Neidle, S. | | 登録日 | 1996-06-26 | | 公開日 | 1997-01-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Designer DNA-binding drugs: the crystal structure of a meta-hydroxy analogue of Hoechst 33258 bound to d(CGCGAATTCGCG)2.
Nucleic Acids Res., 24, 1996
|
|
302D
 
 | | META-HYDROXY ANALOGUE OF HOECHST 33258 ('HYDROXYL IN' CONFORMATION) BOUND TO D(CGCGAATTCGCG)2 | | 分子名称: | 3-[5-[5-(4-METHYL-PIPERAZIN-1-YL)-1H-IMIDAZO[4,5-B]PYRIDIN-2-YL]-BENZIMIDAZOL-2-YL]-PHENOL, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | 著者 | Clark, G.R, Squire, C.J, Gray, E.J, Leupin, W, Neidle, S. | | 登録日 | 1996-06-26 | | 公開日 | 1997-01-20 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Designer DNA-binding drugs: the crystal structure of a meta-hydroxy analogue of Hoechst 33258 bound to d(CGCGAATTCGCG)2.
Nucleic Acids Res., 24, 1996
|
|
1D46
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX:-100 DEGREES C, PIPERAZINE DOWN | | 分子名称: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | 著者 | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | 登録日 | 1991-06-04 | | 公開日 | 1992-04-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
1D43
 
 | | DNA DODECAMER C-G-C-G-A-A-T-T-C-G-C-G/HOECHST 33258 COMPLEX: 0 DEGREES C, PIPERAZINE UP | | 分子名称: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | 著者 | Quintana, J.R, Lipanov, A.A, Dickerson, R.E. | | 登録日 | 1991-06-04 | | 公開日 | 1992-04-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Low-temperature crystallographic analyses of the binding of Hoechst 33258 to the double-helical DNA dodecamer C-G-C-G-A-A-T-T-C-G-C-G.
Biochemistry, 30, 1991
|
|
