7DJ2
 
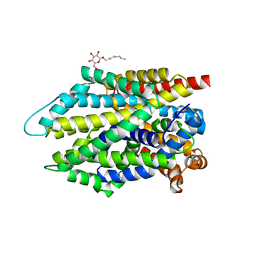 | | Crystal structure of the G26C/E290S mutant of LeuT | | 分子名称: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | 著者 | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | 登録日 | 2020-11-19 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
7DJC
 
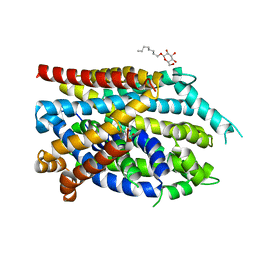 | | Crystal structure of the G26C/Q250A mutant of LeuT | | 分子名称: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION, ... | | 著者 | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | 登録日 | 2020-11-20 | | 公開日 | 2021-04-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
8HZY
 
 | |
8HZV
 
 | |
8HFK
 
 | |
8HFJ
 
 | | Crystal Structure of CbAR mutant (H162F) in complex with NADP+ and a bulky 1,3-cyclodiketone | | 分子名称: | 2-methyl-2-[(4-methylphenyl)methyl]cyclopentane-1,3-dione, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Versicolorin reductase | | 著者 | Hou, X.D, Yin, D.J, Rao, Y.J. | | 登録日 | 2022-11-10 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural analysis of an anthrol reductase inspires enantioselective synthesis of enantiopure hydroxycycloketones and beta-halohydrins.
Nat Commun, 14, 2023
|
|
7Y3Y
 
 | | Crystal structure of BTG13 mutant (T299V) | | 分子名称: | FE (III) ION, GLYCEROL, questin oxidase BTG13 | | 著者 | Hou, X.D, Fu, K, Rao, Y.J. | | 登録日 | 2022-06-13 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y3X
 
 | | Crystal structure of BTG13 mutant (H58F) | | 分子名称: | FE (III) ION, GLYCEROL, Questin oxidase | | 著者 | Hou, X.D, Rao, Y.J. | | 登録日 | 2022-06-13 | | 公開日 | 2022-09-21 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y3W
 
 | |
3BWH
 
 | | Atomic resolution structure of cucurmosin, a novel type 1 RIP from the sarcocarp of Cucurbita moschata | | 分子名称: | 1,2-ETHANEDIOL, PHOSPHATE ION, beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)][alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Chen, L. | | 登録日 | 2008-01-09 | | 公開日 | 2008-10-07 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Atomic resolution structure of cucurmosin, a novel type 1 ribosome-inactivating protein from the sarcocarp of Cucurbita moschata.
J.Struct.Biol., 164, 2008
|
|
7YB1
 
 | |
7YB2
 
 | | Crystal Structure of anthrol reductase (CbAR) in complex with NADP+ and emodin | | 分子名称: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Hou, X.D, Rao, Y.J. | | 登録日 | 2022-06-28 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural analysis of an anthrol reductase inspires enantioselective synthesis of enantiopure hydroxycycloketones and beta-halohydrins.
Nat Commun, 14, 2023
|
|
8K4F
 
 | | DHODH in complex with compound A0 | | 分子名称: | 5-cyclopropyl-2-[1-[(2-fluorophenyl)methyl]pyrazolo[3,4-b]pyridin-3-yl]pyrimidin-4-amine, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, ACETATE ION, ... | | 著者 | Jian, L, Sun, Q. | | 登録日 | 2023-07-18 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Discovery and Optimization of Novel h DHODH Inhibitors for the Treatment of Inflammatory Bowel Disease.
J.Med.Chem., 66, 2023
|
|
7MF1
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing antibody 47D1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 47D1 Fab heavy chain, ... | | 著者 | Yuan, M, Zhu, X, Wilson, I.A. | | 登録日 | 2021-04-08 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.092 Å) | | 主引用文献 | Diverse immunoglobulin gene usage and convergent epitope targeting in neutralizing antibody responses to SARS-CoV-2.
Cell Rep, 35, 2021
|
|
6D39
 
 | |
6D38
 
 | |
8SMK
 
 | | hPAD4 bound to Activating Fab hA362 | | 分子名称: | Activating Fab 362 heavy chain, Activating Fab 362 light chain, CALCIUM ION, ... | | 著者 | Maker, A, Verba, K.A. | | 登録日 | 2023-04-26 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Nat.Chem.Biol., 20, 2024
|
|
8SML
 
 | | hPAD4 bound to inhibitory Fab hI365 | | 分子名称: | CALCIUM ION, Fab hI365 heavy chain, Fab hI365 light chain, ... | | 著者 | Maker, A, Verba, K.A. | | 登録日 | 2023-04-26 | | 公開日 | 2024-03-06 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Antibody discovery identifies regulatory mechanisms of protein arginine deiminase 4.
Nat.Chem.Biol., 20, 2024
|
|
8HCO
 
 | | Substrate-engaged TOM complex from yeast | | 分子名称: | Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, ... | | 著者 | Zhou, X.Y, Yang, Y.Q, Wang, G.P, Wang, S.S. | | 登録日 | 2022-11-02 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-12-27 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Molecular pathway of mitochondrial preprotein import through the TOM-TIM23 supercomplex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7C2Q
 
 | | The crystal structure of COVID-19 main protease in the apo state | | 分子名称: | 3C-like proteinase | | 著者 | Zhou, X.L, Zhong, F.L, Lin, C, Hu, X.H, Zhou, H, Wang, Q.S, Li, j, Zhang, J. | | 登録日 | 2020-05-08 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structure of SARS-CoV-2 main protease in the apo state.
Sci China Life Sci, 64, 2021
|
|
6L6Z
 
 | |
6LFG
 
 | |
7WJ4
 
 | |
7WIZ
 
 | |
4R5P
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and a nucleoside triphosphate mimic alpha-carboxy nucleoside phosphonate inhibitor | | 分子名称: | 5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', 5'-D(*TP*GP*GP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3', HIV-1 reverse transcriptase, ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2014-08-21 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.894 Å) | | 主引用文献 | Alpha-carboxy nucleoside phosphonates as universal nucleoside triphosphate mimics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
