6XX2
 
 | |
6S7N
 
 | |
6SYC
 
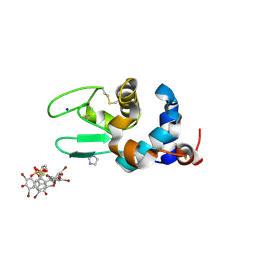 | | Crystal structure of the lysozyme in presence of bromophenol blue at pH 6.5 | | 分子名称: | CHLORIDE ION, IMIDAZOLE, Lysozyme, ... | | 著者 | Camara-Artigas, A, Plaza-Garrido, M, Salinas-Garcia, M.C. | | 登録日 | 2019-09-27 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Lysozyme crystals dyed with bromophenol blue: where has the dye gone?
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6SYD
 
 | |
6SYE
 
 | |
4OMO
 
 | |
4OML
 
 | |
4OMN
 
 | |
4QT7
 
 | |
4OMP
 
 | |
4R61
 
 | |
4REX
 
 | |
4JZ4
 
 | | Crystal structure of chicken c-Src-SH3 domain: monomeric form | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, NICKEL (II) ION, Proto-oncogene tyrosine-protein kinase Src | | 著者 | Camara-Artigas, A. | | 登録日 | 2013-04-02 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Electrostatic Effects in the Folding of the SH3 Domain of the c-Src Tyrosine Kinase: pH-Dependence in 3D-Domain Swapping and Amyloid Formation.
Plos One, 9, 2014
|
|
4JZ3
 
 | |
6F9Y
 
 | |
6F1L
 
 | |
6F1P
 
 | |
6F9Z
 
 | |
6F1M
 
 | |
6F1O
 
 | |
6F1R
 
 | |
6FA0
 
 | |
6F9X
 
 | |
1U87
 
 | |
8QPD
 
 | |
