5WCH
 
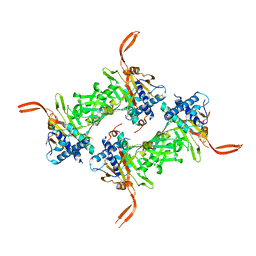 | | Crystal structure of the catalytic domain of human USP9X | | 分子名称: | Probable ubiquitin carboxyl-terminal hydrolase FAF-X, UNKNOWN ATOM OR ION, ZINC ION | | 著者 | Dong, A, Zhang, Q, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | 登録日 | 2017-06-30 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and activity-based labeling reveal the mechanisms for linkage-specific substrate recognition by deubiquitinase USP9X.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6KJX
 
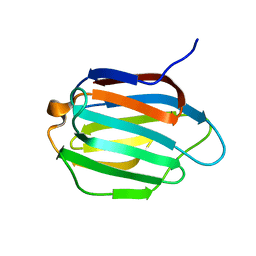 | | Galectin-13 variant C138S | | 分子名称: | Galactoside-binding soluble lectin 13 | | 著者 | Su, J. | | 登録日 | 2019-07-23 | | 公開日 | 2019-10-16 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
7VOK
 
 | |
4NK5
 
 | |
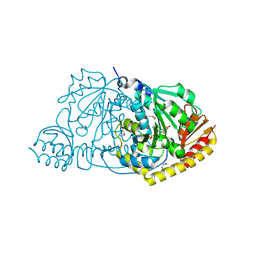
1FG3
 
 | |
6XKB
 
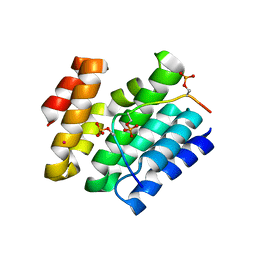 | | Crystal structure of SR-related and CTD-associated factor 4(SCAF4-CID)with peptide S2,S5p-CTD | | 分子名称: | S2,S5p-CTD peptide, SR-related and CTD-associated factor 4, UNKNOWN ATOM OR ION | | 著者 | Zhou, M.Q, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2020-06-26 | | 公開日 | 2021-01-20 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the recognition of the S2, S5-phosphorylated RNA polymerase II CTD by the mRNA anti-terminator protein hSCAF4.
Febs Lett., 596, 2022
|
|
4N9G
 
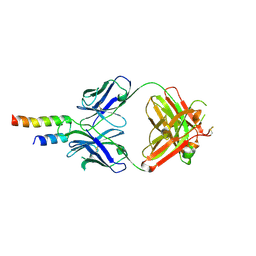 | |
7E7Y
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.16 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | 分子名称: | 368-2 H, 368-2 L, 604 H, ... | | 著者 | Du, S, Xiao, J, Zhang, Z. | | 登録日 | 2021-03-01 | | 公開日 | 2021-06-09 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E7X
 
 | |
7E86
 
 | |
7E88
 
 | |
3JBZ
 
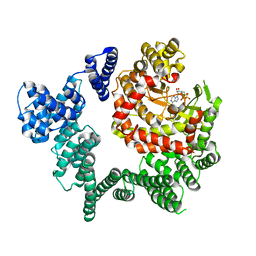 | |
6WS5
 
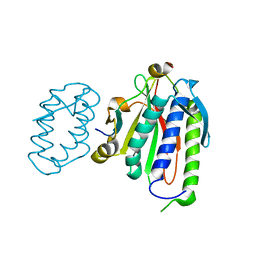 | | Rational drug design of phenazopyridine derivatives as novel inhibitors of Rev1-CT | | 分子名称: | 3-[(Z)-(2,3-difluorophenyl)diazenyl]pyridine-2,6-diamine, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | 著者 | McPherson, K.S, Korzhnev, D.M. | | 登録日 | 2020-04-30 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.472 Å) | | 主引用文献 | Structure-Based Drug Design of Phenazopyridine Derivatives as Inhibitors of Rev1 Interactions in Translesion Synthesis.
Chemmedchem, 16, 2021
|
|
1HN0
 
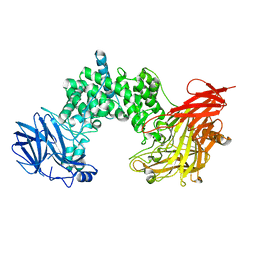 | |
1HSI
 
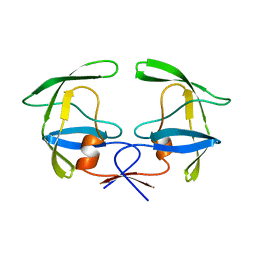 | |
1HSG
 
 | |
1F4Q
 
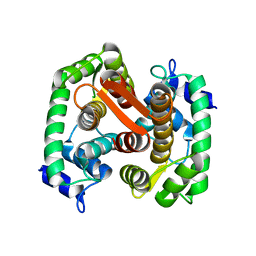 | | CRYSTAL STRUCTURE OF APO GRANCALCIN | | 分子名称: | GRANCALCIN | | 著者 | Jia, J, Han, Q, Borregaard, N, Lollike, K, Cygler, M. | | 登録日 | 2000-06-08 | | 公開日 | 2000-09-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of human grancalcin, a member of the penta-EF-hand protein family.
J.Mol.Biol., 300, 2000
|
|
1F4O
 
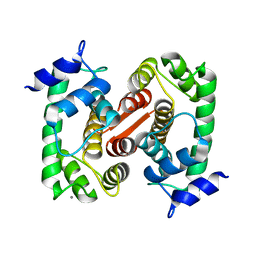 | | CRYSTAL STRUCTURE OF GRANCALCIN WITH BOUND CALCIUM | | 分子名称: | CALCIUM ION, GRANCALCIN | | 著者 | Jia, J, Han, Q, Borregaard, N, Lollike, K, Cygler, M. | | 登録日 | 2000-06-08 | | 公開日 | 2000-09-27 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of human grancalcin, a member of the penta-EF-hand protein family.
J.Mol.Biol., 300, 2000
|
|
1IM6
 
 | |
1FG7
 
 | |
1F9H
 
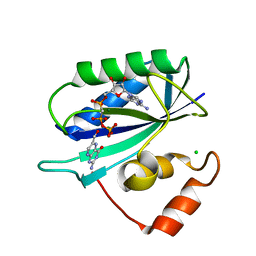 | | CRYSTAL STRUCTURE OF THE TERNARY COMPLEX OF E. COLI HPPK(R92A) WITH MGAMPCPP AND 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN AT 1.50 ANGSTROM RESOLUTION | | 分子名称: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE, CHLORIDE ION, ... | | 著者 | Blaszczyk, J, Ji, X. | | 登録日 | 2000-07-10 | | 公開日 | 2003-04-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Dynamic Roles of Arginine Residues 82 and 92 of Escherichia coli
6-Hydroxymethyl-7,8-dihydropterin Pyrophosphokinase: Crystallographic
Studies
Biochemistry, 42, 2003
|
|
8H7Z
 
 | |
8H7L
 
 | | Cryo-EM Structure of SARS-CoV-2 BA.2 Spike protein in complex with BA7535 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7535 fab heavt chain, ... | | 著者 | Liu, Z, Yan, A, Gao, Y. | | 登録日 | 2022-10-20 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.44 Å) | | 主引用文献 | Identification of a highly conserved neutralizing epitope within the RBD region of diverse SARS-CoV-2 variants.
Nat Commun, 15, 2024
|
|
