7DD4
 
 | |
2Q33
 
 | | Crystal structure of all-D monellin at 1.8 A resolution | | 分子名称: | D-MONELLIN CHAIN A, D-MONELLIN CHAIN B | | 著者 | Hung, L.-W, Kohmura, M, Ariyoshi, Y, Kim, S.-H. | | 登録日 | 2007-05-29 | | 公開日 | 2007-11-13 | | 最終更新日 | 2017-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of an Enantiomeric Protein, D-Monellin at 1.8 A Resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6K5W
 
 | |
6L4P
 
 | | Crystal structure of the complex between the axonemal outer-arm dynein light chain-1 and microtubule binding domain of gamma heavy chain | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Dynein light chain 1, axonemal, ... | | 著者 | Toda, A, Nishikawa, Y, Tanaka, H, Yagi, T, Kurisu, G. | | 登録日 | 2019-10-19 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.703 Å) | | 主引用文献 | The complex of outer-arm dynein light chain-1 and the microtubule-binding domain of the gamma heavy chain shows how axonemal dynein tunes ciliary beating.
J.Biol.Chem., 295, 2020
|
|
6L1X
 
 | | Quinol-dependent nitric oxide reductase (qNOR) from Neisseria meningitidis in the monomeric oxidized state with zinc complex. | | 分子名称: | CALCIUM ION, FE (III) ION, Nitric-oxide reductase, ... | | 著者 | Jamali, M.M.A, Antonyuk, S.V, Tosha, T, Muramoto, K, Hasnain, S.S, Shiro, Y. | | 登録日 | 2019-10-01 | | 公開日 | 2020-04-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | The active form of quinol-dependent nitric oxide reductase fromNeisseria meningitidisis a dimer.
Iucrj, 7, 2020
|
|
1WPK
 
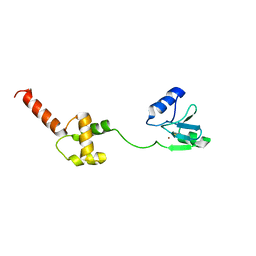 | | Methylated Form of N-terminal Transcriptional Regulator Domain of Escherichia Coli Ada Protein | | 分子名称: | ADA regulatory protein, ZINC ION | | 著者 | Takinowaki, H, Matsuda, Y, Yoshida, T, Kobayashi, Y, Ohkubo, T. | | 登録日 | 2004-09-07 | | 公開日 | 2005-09-13 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of the methylated form of the N-terminal 16-kDa domain of Escherichia coli Ada protein
Protein Sci., 15, 2006
|
|
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-07-12 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | 著者 | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | 登録日 | 2022-07-13 | | 公開日 | 2023-07-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
2AI5
 
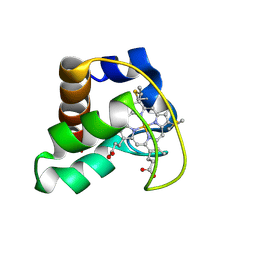 | | Solution Structure of Cytochrome C552, determined by Distributed Computing Implementation for NMR data | | 分子名称: | Cytochrome c-552, HEME C | | 著者 | Nakamura, S, Ichiki, S.I, Takashima, H, Uchiyama, S, Hasegawa, J, Kobayashi, Y, Sambongi, Y, Ohkubo, T. | | 登録日 | 2005-07-29 | | 公開日 | 2006-05-23 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of Cytochrome c552 from a Moderate Thermophilic Bacterium, Hydrogenophilus thermoluteolus: Comparative Study on the Thermostability of Cytochrome c
Biochemistry, 45, 2006
|
|
7E15
 
 | |
1UHZ
 
 | | Solution structure of dsRNA binding domain in Staufen homolog 2 | | 分子名称: | staufen (RNA binding protein) homolog 2 | | 著者 | He, F, Muto, Y, Obayashi, N, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Koboyashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-07-14 | | 公開日 | 2004-08-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of dsRNA binding domain in Staufen homolog 2
To be Published
|
|
2E5D
 
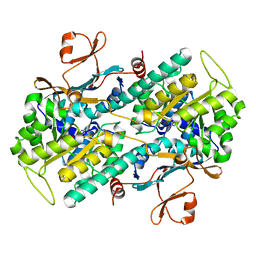 | | Crystal structure of Human NMPRTase complexed with nicotinamide | | 分子名称: | NICOTINAMIDE, Nicotinamide phosphoribosyltransferase | | 著者 | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | 登録日 | 2006-12-20 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
2E5C
 
 | | Crystal structure of Human NMPRTase complexed with 5'-phosphoribosyl-1'-pyrophosphate | | 分子名称: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, Nicotinamide phosphoribosyltransferase | | 著者 | Takahashi, R, Nakamura, S, Kobayashi, Y, Ohkubo, T. | | 登録日 | 2006-12-20 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase
J.Biochem., 147, 2010
|
|
1V55
 
 | | Bovine heart cytochrome c oxidase at the fully reduced state | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | 著者 | Tsukihara, T, Shimokata, K, Katayama, Y, Shimada, H, Muramoto, K, Aoyama, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yao, M, Ishimura, Y, Yoshikawa, S. | | 登録日 | 2003-11-21 | | 公開日 | 2003-12-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
1V54
 
 | | Bovine heart cytochrome c oxidase at the fully oxidized state | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | 著者 | Tsukihara, T, Shimokata, K, Katayama, Y, Shimada, H, Muramoto, K, Aoyama, H, Mochizuki, M, Shinzawa-Itoh, K, Yamashita, E, Yao, M, Ishimura, Y, Yoshikawa, S. | | 登録日 | 2003-11-21 | | 公開日 | 2003-12-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
2E5B
 
 | |
3B2C
 
 | | Crystal structure of the collagen triple helix model [{PRO-HYP(R)-GLY}4-{HYP(S)-Pro-GLY}2-{PRO-HYP(R)-GLY}4]3 | | 分子名称: | Collagen-like peptide | | 著者 | Motooka, D, Kawahara, K, Nakamura, S, Doi, M, Nishi, Y, Nishiuchi, Y, Nakazawa, T, Yoshida, T, Ohkubo, T, Kobayashi, Y, Kang, Y.K, Uchiyama, S. | | 登録日 | 2011-07-26 | | 公開日 | 2012-04-04 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | The triple helical structure and stability of collagen model peptide with 4(S)-hydroxyprolyl-pro-gly units
Biopolymers, 98, 2011
|
|
1UHW
 
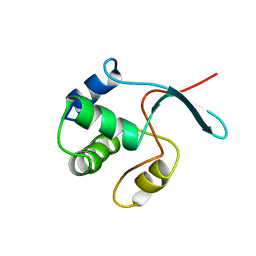 | | Solution structure of the DEP domain of mouse pleckstrin | | 分子名称: | Pleckstrin | | 著者 | Inoue, K, Yoshida, M, Hatta, R, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-07-11 | | 公開日 | 2004-01-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the DEP domain of mouse pleckstrin
To be Published
|
|
3CV9
 
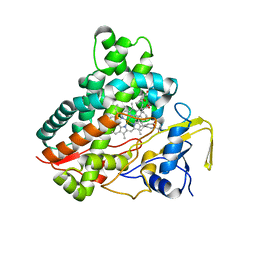 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R73A/R84A mutant) in complex with 1alpha,25-dihydroxyvitamin D3 | | 分子名称: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Hayashi, K, Sugimoto, H, Shinkyo, R, Yamada, M, Ikeda, S, Ikushiro, S, Kamakura, M, Shiro, Y, Sakaki, T. | | 登録日 | 2008-04-18 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1
Biochemistry, 47, 2008
|
|
3CV8
 
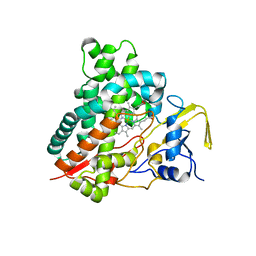 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84F mutant) | | 分子名称: | Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Hayashi, K, Sugimoto, H, Shinkyo, R, Yamada, M, Ikeda, S, Ikushiro, S, Kamakura, M, Shiro, Y, Sakaki, T. | | 登録日 | 2008-04-18 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1
Biochemistry, 47, 2008
|
|
7EAQ
 
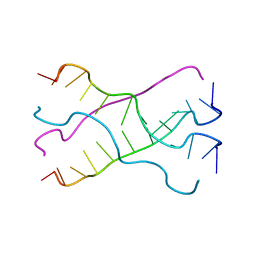 | |
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | 分子名称: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | 著者 | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | 登録日 | 2021-10-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7F4Z
 
 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Heat shock 70 kDa protein 1B, ... | | 著者 | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | 登録日 | 2021-06-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
7F50
 
 | | X-ray crystal structure of Y149A mutated Hsp72-NBD in complex with AMPPnP | | 分子名称: | CHLORIDE ION, Heat shock 70 kDa protein 1B, MAGNESIUM ION, ... | | 著者 | Yokoyama, T, Fujii, S, Nabeshima, Y, Mizuguchi, M. | | 登録日 | 2021-06-21 | | 公開日 | 2022-06-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.703 Å) | | 主引用文献 | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | 登録日 | 2021-10-18 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
