3ZQ1
 
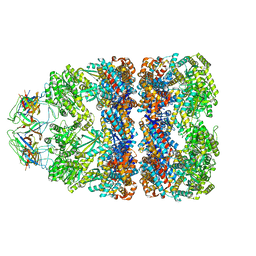 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | 分子名称: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | 登録日 | 2013-03-04 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (15.9 Å) | | 主引用文献 | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
6SUH
 
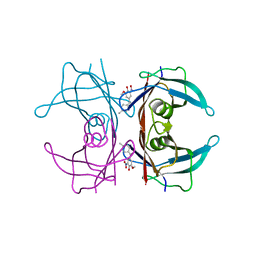 | | Crystal structure of human transthyretin in complex with 3-O-methyltolcapone, a tolcapone analogue | | 分子名称: | 3-O-methyltolcapone, Transthyretin | | 著者 | Loconte, V, Cianci, M, Menozzi, I, Sbravati, D, Sansone, F, Casnati, A, Berni, R. | | 登録日 | 2019-09-14 | | 公開日 | 2020-08-05 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Interactions of tolcapone analogues as stabilizers of the amyloidogenic protein transthyretin.
Bioorg.Chem., 103, 2020
|
|
6SUP
 
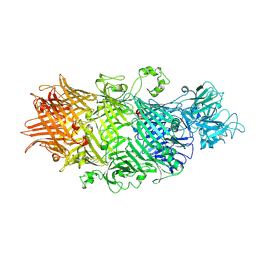 | | Crystal Structure of TcdB2-TccC3-Cdc42 | | 分子名称: | MAGNESIUM ION, TcdB2,TccC3,Cell division control protein 42 homolog | | 著者 | Roderer, D, Schubert, E, Sitsel, O, Raunser, S. | | 登録日 | 2019-09-16 | | 公開日 | 2019-12-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Towards the application of Tc toxins as a universal protein translocation system.
Nat Commun, 10, 2019
|
|
2O03
 
 | |
7ZHC
 
 | | Moss spermine/spermidine acetyl transferase (PpSSAT) in complex with AcetylCoA and polyethylen glycol | | 分子名称: | ACETYL COENZYME *A, GLYCEROL, N-acetyltransferase domain-containing protein, ... | | 著者 | Morera, S, Kopecny, D, Vigouroux, A. | | 登録日 | 2022-04-06 | | 公開日 | 2023-03-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.819 Å) | | 主引用文献 | Biochemical and structural basis of polyamine, lysine and ornithine acetylation catalyzed by spermine/spermidine N-acetyl transferase in moss and maize.
Plant J., 114, 2023
|
|
3ZQ0
 
 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | 分子名称: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | 登録日 | 2013-03-04 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (9.2 Å) | | 主引用文献 | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
2O2D
 
 | | Crystal structure of phosphoglucose isomerase from Trypanosoma brucei complexed with citrate | | 分子名称: | CITRIC ACID, GLYCEROL, Glucose-6-phosphate isomerase, ... | | 著者 | Arsenieva, D, Mazock, G.H, Appavu, B.L, Jeffery, C.J. | | 登録日 | 2006-11-29 | | 公開日 | 2007-11-13 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of phosphoglucose isomerase from Trypanosoma brucei complexed with glucose-6-phosphate at 1.6 A resolution
Proteins, 74, 2008
|
|
3ZL2
 
 | | A thiazolyl-mannoside bound to FimH, orthorhombic space group | | 分子名称: | N-{5-[(1R)-1-hydroxyethyl]-1,3-thiazol-2-yl}-alpha-D-mannopyranosylamine, PROTEIN FIMH | | 著者 | Brument, S, Sivignon, A, Dumych, T.I, Moreau, N, Roos, G, Guerardel, Y, Chalopin, T, Deniaud, D, Bilyy, R.O, Darfeuille-Michaud, A, Bouckaert, J, Gouin, S.G. | | 登録日 | 2013-01-27 | | 公開日 | 2013-07-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.251 Å) | | 主引用文献 | Thiazolylaminomannosides as Potent Antiadhesives of Type 1 Piliated Escherichia Coli Isolated from Crohn'S Disease Patients.
J.Med.Chem., 56, 2013
|
|
3K49
 
 | | Puf3 RNA binding domain bound to Cox17 RNA 3' UTR recognition sequence site B | | 分子名称: | CITRIC ACID, RNA (5'-R(*CP*CP*UP*GP*UP*AP*AP*AP*UP*A)-3'), mRNA-binding protein PUF3 | | 著者 | Zhu, D, Stumpf, C.R, Krahn, J.M, Wickens, M, Hall, T.M.T. | | 登録日 | 2009-10-05 | | 公開日 | 2009-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A 5' cytosine binding pocket in Puf3p specifies regulation of mitochondrial mRNAs.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2O3V
 
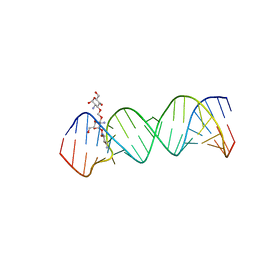 | | Crystal Structure of the Homo sapiens Cytoplasmic Ribosomal Decoding Site complexed with paromamine derivative NB33 | | 分子名称: | (2S,3R,4R,5S,6R)-3-AMINO-4-({[(2S,3R,4R,5S,6R)-3-AMINO-2-{[(1R,2R,3S,4R,6S)-4,6-DIAMINO-2,3-DIHYDROXYCYCLOHEXYL]OXY}-5-HYDROXY-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-4-YL]OXY}METHOXY)-6-(HYDROXYMETHYL)TETRAHYDRO-2H-PYRAN-2,5-DIOL, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*CP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | 著者 | Kondo, J, Hainrichson, M, Nudelman, I, Shallom-Shezifi, D, Baasov, T, Westhof, E. | | 登録日 | 2006-12-02 | | 公開日 | 2007-11-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Differential Selectivity of Natural and Synthetic Aminoglycosides towards the Eukaryotic and Prokaryotic Decoding A Sites.
Chembiochem, 8, 2007
|
|
6SKO
 
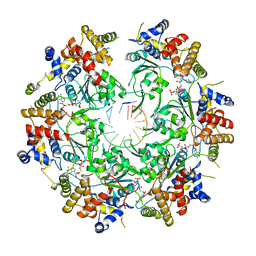 | | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork - conformation 2 MCM CTD:ssDNA | | 分子名称: | DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, DNA replication licensing factor MCM4, ... | | 著者 | Yeeles, J, Baretic, D, Jenkyn-Bedford, M. | | 登録日 | 2019-08-16 | | 公開日 | 2020-05-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM Structure of the Fork Protection Complex Bound to CMG at a Replication Fork.
Mol.Cell, 78, 2020
|
|
2O2N
 
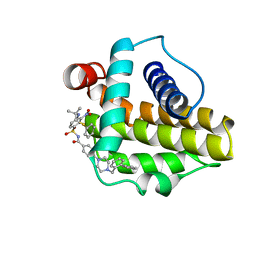 | | Solution structure of the anti-apoptotic protein Bcl-xL in complex with an acyl-sulfonamide-based ligand | | 分子名称: | 4-[4-(BIPHENYL-2-YLMETHYL)PIPERAZIN-1-YL]-N-[(4-{[1,1-DIMETHYL-2-(PHENYLTHIO)ETHYL]AMINO}-3-NITROPHENYL)SULFONYL]BENZAMIDE, Apoptosis regulator Bcl-X | | 著者 | Bruncko, M, Oost, T.K, Belli, B.A, Ding, H, Joseph, M.K, Kunzer, A, Martineau, D, McClellan, W.J, Mitten, M, Ng, S.C, Nimmer, P.M, Oltersdorf, T, Park, C.M, Petros, A.M, Shoemaker, A.R, Song, X, Wang, X, Wendt, M.D, Zhang, H, Fesik, S.W, Rosenberg, S.H, Elmore, S.W. | | 登録日 | 2006-11-30 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Studies Leading to Potent, Dual Inhibitors of Bcl-2 and Bcl-xL.
J.Med.Chem., 50, 2007
|
|
6SYM
 
 | |
8BH5
 
 | | SARS-CoV-2 BA.2.12.1 RBD in complex with Beta-27 Fab and C1 nanobody | | 分子名称: | Beta-27 heavy chain, Beta-27 light chain, GLYCEROL, ... | | 著者 | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | 登録日 | 2022-10-29 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Humoral responses against SARS-CoV-2 Omicron BA.2.11, BA.2.12.1 and BA.2.13 from vaccine and BA.1 serum.
Cell Discov, 8, 2022
|
|
6SZL
 
 | | Crystal structure of YTHDC1 with fragment 7 (DHU_DC1_021) | | 分子名称: | 6-phenyl-1~{H}-pyrimidine-2,4-dione, SULFATE ION, YTH domain-containing protein 1 | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
3J4J
 
 | | Model of full-length T. thermophilus Translation Initiation Factor 2 refined against its cryo-EM density from a 30S Initiation Complex map | | 分子名称: | Translation initiation factor IF-2 | | 著者 | Simonetti, A, Marzi, S, Billas, I.M.L, Tsai, A, Fabbretti, A, Myasnikov, A, Roblin, P, Vaiana, A.C, Hazemann, I, Eiler, D, Steitz, T.A, Puglisi, J.D, Gualerzi, C.O, Klaholz, B.P. | | 登録日 | 2013-08-26 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (11.5 Å) | | 主引用文献 | Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6SZQ
 
 | | Crystal structure of human DDAH-1 | | 分子名称: | N(G),N(G)-dimethylarginine dimethylaminohydrolase 1 | | 著者 | Hennig, S, Vetter, I.R, Schade, D. | | 登録日 | 2019-10-02 | | 公開日 | 2019-12-25 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.412 Å) | | 主引用文献 | Discovery ofN-(4-Aminobutyl)-N'-(2-methoxyethyl)guanidine as the First Selective, Nonamino Acid, Catalytic Site Inhibitor of Human Dimethylarginine Dimethylaminohydrolase-1 (hDDAH-1).
J.Med.Chem., 63, 2020
|
|
6T09
 
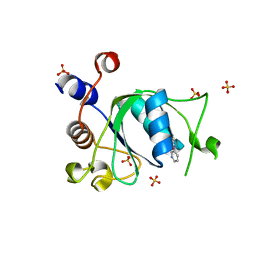 | | Crystal structure of YTHDC1 with fragment 24 (PSI_DC1_003) | | 分子名称: | SULFATE ION, YTHDC1, ~{N}-pyridin-3-ylethanamide | | 著者 | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | 登録日 | 2019-10-02 | | 公開日 | 2020-03-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
1CP2
 
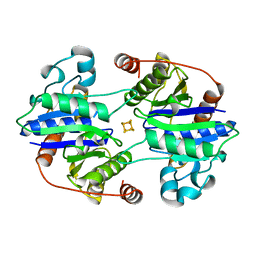 | | NITROGENASE IRON PROTEIN FROM CLOSTRIDIUM PASTEURIANUM | | 分子名称: | IRON/SULFUR CLUSTER, NITROGENASE IRON PROTEIN | | 著者 | Schlessman, J.L, Woo, D, Joshua-Tor, L, Howard, J.B, Rees, D.C. | | 登録日 | 1998-05-11 | | 公開日 | 1998-11-04 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Conformational variability in structures of the nitrogenase iron proteins from Azotobacter vinelandii and Clostridium pasteurianum.
J.Mol.Biol., 280, 1998
|
|
6SGS
 
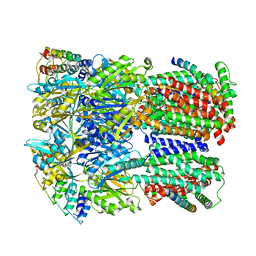 | | Cryo-EM structure of Escherichia coli AcrBZ and DARPin in Saposin A-nanodisc | | 分子名称: | DARPin, Multidrug efflux pump accessory protein AcrZ, Multidrug efflux pump subunit AcrB | | 著者 | Szewczak-Harris, A, Du, D, Newman, C, Neuberger, A, Luisi, B.F. | | 登録日 | 2019-08-05 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment.
Structure, 28, 2020
|
|
3J6K
 
 | |
6SPB
 
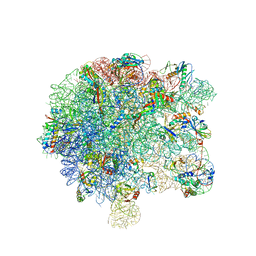 | | Pseudomonas aeruginosa 50s ribosome from a clinical isolate with a mutation in uL6 | | 分子名称: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | 著者 | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | 登録日 | 2019-09-01 | | 公開日 | 2019-10-16 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (2.82 Å) | | 主引用文献 | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8B8A
 
 | | Multimerization domain of borna disease virus 1 phosphoprotein | | 分子名称: | Phosphoprotein | | 著者 | Tarbouriech, N, Legrand, P, Bourhis, J.M, Chenavier, F, Freslon, L, Kawasaki, J, Horie, M, Tomonaga, K, Bachiri, K, Coyaud, E, Gonzalez-Dunia, D, Ruigrok, R.W.H, Crepin, T. | | 登録日 | 2022-10-04 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Borna Disease Virus 1 Phosphoprotein Forms a Tetramer and Interacts with Host Factors Involved in DNA Double-Strand Break Repair and mRNA Processing.
Viruses, 14, 2022
|
|
2O7P
 
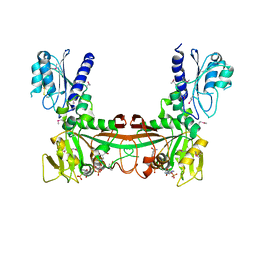 | | The crystal structure of RibD from Escherichia coli in complex with the oxidised NADP+ cofactor in the active site of the reductase domain | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Riboflavin biosynthesis protein ribD | | 著者 | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | 登録日 | 2006-12-11 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
2OBC
 
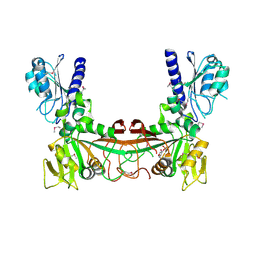 | | The crystal structure of RibD from Escherichia coli in complex with a substrate analogue, ribose 5-phosphate (beta form), bound to the active site of the reductase domain | | 分子名称: | 5-O-phosphono-beta-D-ribofuranose, Riboflavin biosynthesis protein ribD | | 著者 | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | 登録日 | 2006-12-18 | | 公開日 | 2007-02-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
