2ATM
 
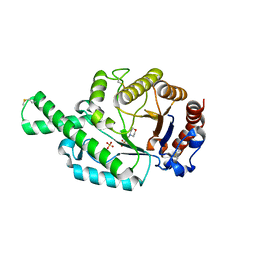 | | Crystal structure of the recombinant allergen Ves v 2 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Hyaluronoglucosaminidase, SULFATE ION | | 著者 | Skov, L.K, Seppala, U, Coen, J.J.F, Crickmore, N, King, T.P, Monsalve, R, Kastrup, J.S, Spangfort, M.D, Gajhede, M. | | 登録日 | 2005-08-25 | | 公開日 | 2006-05-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of recombinant Ves v 2 at 2.0 Angstrom resolution: structural analysis of an allergenic hyaluronidase from wasp venom.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
6WHL
 
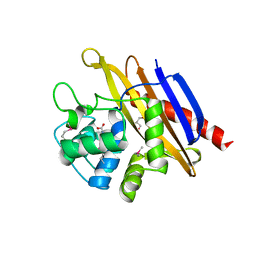 | |
1QO7
 
 | | Structure of Aspergillus niger epoxide hydrolase | | 分子名称: | EPOXIDE HYDROLASE | | 著者 | Zou, J.-Y, Hallberg, B.M, Bergfors, T, Oesch, F, Arand, M, Mowbray, S.L, Jones, T.A. | | 登録日 | 1999-11-04 | | 公開日 | 2000-02-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of Aspergillus Niger Epoxide Hydrolase at 1.8A Resolution: Implications for the Structure and Function of the Mammalian Microsomal Class of Epoxide Hydrolases
Structure, 8, 2000
|
|
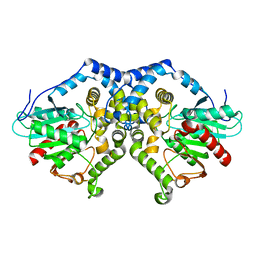
2AVW
 
 | | Crystal structure of monoclinic form of streptococcus Mac-1 | | 分子名称: | GLYCEROL, IgG-degrading protease, SULFATE ION | | 著者 | Agniswamy, J, Nagiec, M.J, Liu, M, Schuck, P, Musser, J.M, Sun, P.D. | | 登録日 | 2005-08-30 | | 公開日 | 2006-02-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of group a streptococcus mac-1: insight into dimer-mediated specificity for recognition of human IgG.
Structure, 14, 2006
|
|
1R69
 
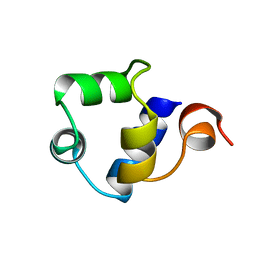 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | 分子名称: | REPRESSOR PROTEIN CI | | 著者 | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | 登録日 | 1988-12-08 | | 公開日 | 1989-10-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
2AYO
 
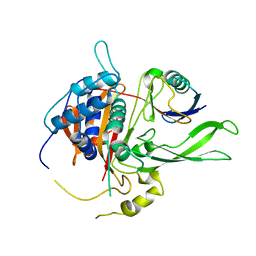 | | Structure of USP14 bound to ubquitin aldehyde | | 分子名称: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 14 | | 著者 | Hu, M, Li, P, Jeffrey, P.D, Shi, Y. | | 登録日 | 2005-09-07 | | 公開日 | 2005-10-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14.
Embo J., 24, 2005
|
|
6WTZ
 
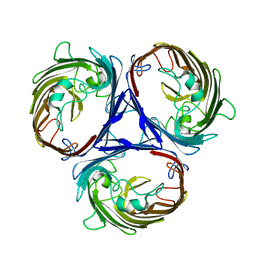 | |
6WVN
 
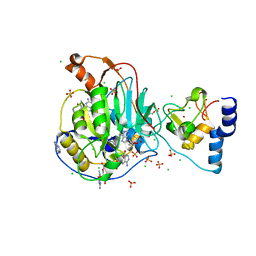 | | Crystal Structure of Nsp16-Nsp10 from SARS-CoV-2 in Complex with 7-methyl-GpppA and S-Adenosylmethionine. | | 分子名称: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, ADENINE, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-06 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | High-resolution structures of the SARS-CoV-2 2'- O -methyltransferase reveal strategies for structure-based inhibitor design.
Sci.Signal., 13, 2020
|
|
1R5H
 
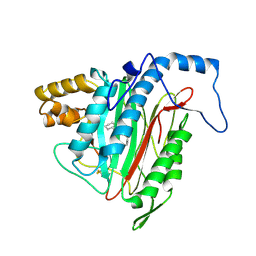 | | Crystal Structure of MetAP2 complexed with A320282 | | 分子名称: | MANGANESE (II) ION, Methionine aminopeptidase 2, N'-(2S,3R)-3-AMINO-4-CYCLOHEXYL-2-HYDROXY-BUTANO-N-(4-METHYLPHENYL)HYDRAZIDE | | 著者 | Sheppard, G.S, Wang, J, Kawai, M, BaMaung, N.Y, Craig, R.A, Erickson, S.A, Lynch, L, Patel, J, Yang, F, Searle, X.B, Lou, P, Park, C, Kim, K.H, Henkin, J, Lesniewski, R. | | 登録日 | 2003-10-10 | | 公開日 | 2004-10-12 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | 3-Amino-2-hydroxyamides and related compounds as inhibitors of methionine aminopeptidase-2.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2ANE
 
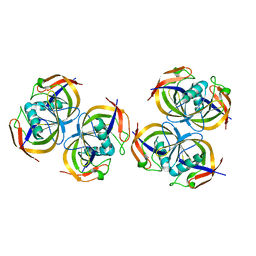 | | Crystal structure of N-terminal domain of E.Coli Lon Protease | | 分子名称: | ATP-dependent protease La | | 著者 | Li, M, Rasulova, F, Melnikov, E.E, Rotanova, T.V, Gustchina, A, Maurizi, M.R, Wlodawer, A. | | 登録日 | 2005-08-11 | | 公開日 | 2005-11-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Crystal structure of the N-terminal domain of E. coli Lon protease.
Protein Sci., 14, 2005
|
|
1R5S
 
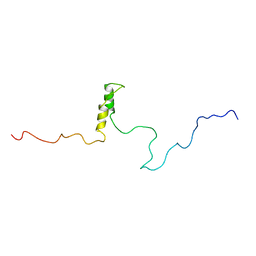 | | Connexin 43 Carboxyl Terminal Domain | | 分子名称: | Gap junction alpha-1 protein | | 著者 | Sorgen, P.L, Duffy, H.S, Mario, D, Sahoo, P, Coombs, W, Delmar, M, Spray, D.C. | | 登録日 | 2003-10-13 | | 公開日 | 2004-10-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1
J.Biol.Chem., 279, 2004
|
|
2AO7
 
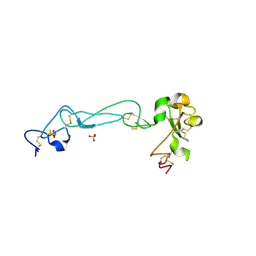 | | Adam10 Disintegrin and cysteine- rich domain | | 分子名称: | ADAM 10, SULFATE ION | | 著者 | Janes, P.W, Saha, N, Barton, W.A, Kolev, M.V, Wimmer-Kleikamp, S.H, Nievergall, E, Blobel, C.P, Himanen, J.-P, Lackmann, M, Nikolov, D.B. | | 登録日 | 2005-08-12 | | 公開日 | 2006-08-08 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans.
Cell(Cambridge,Mass.), 123, 2005
|
|
6X0L
 
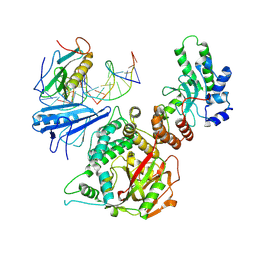 | |
2APQ
 
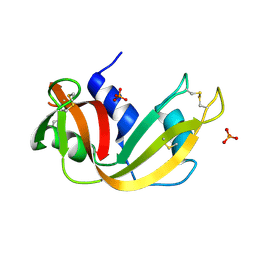 | | Crystal Structure of an Active Site Mutant of Bovine Pancreatic Ribonuclease A (H119A-RNase A) with a 10-Glutamine expansion in the C-terminal hinge-loop. | | 分子名称: | PHOSPHATE ION, Ribonuclease | | 著者 | Sambashivan, S, Liu, Y, Sawaya, M.R, Gingery, M, Eisenberg, D. | | 登録日 | 2005-08-16 | | 公開日 | 2005-09-13 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure.
Nature, 437, 2005
|
|
1R7H
 
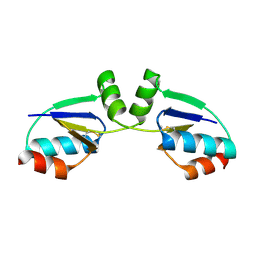 | |
6X0M
 
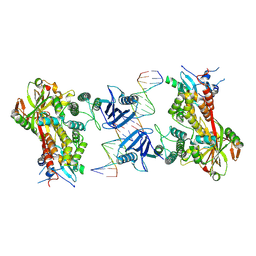 | |
2AL7
 
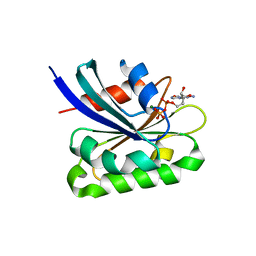 | | Structure Of Human ADP-Ribosylation Factor-Like 10C | | 分子名称: | ADP-ribosylation factor-like 10C, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | 著者 | Ismail, S, Dimov, S, Atanassova, A, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | 登録日 | 2005-08-04 | | 公開日 | 2005-08-23 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | GTP-like conformation of GDP-bound ARL10C GTPase
To be Published
|
|
6X1L
 
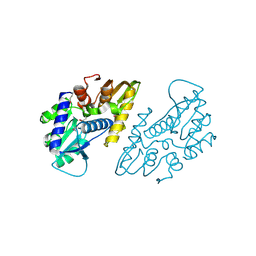 | | The crystal structure of a functional uncharacterized protein KP1_0663 from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | | 分子名称: | WbbZ protein | | 著者 | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-19 | | 公開日 | 2020-06-03 | | 最終更新日 | 2023-06-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6X42
 
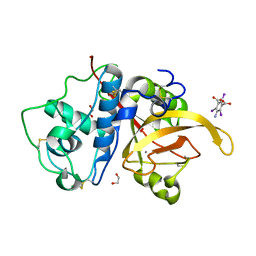 | | High Resolution Crystal Structure Analysis of SERA5E from plasmodium falciparum | | 分子名称: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CALCIUM ION, ... | | 著者 | Clarke, O.B, Smith, N.A, Lee, M, Smith, B.J. | | 登録日 | 2020-05-21 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structure of the Plasmodium falciparum PfSERA5 pseudo-zymogen.
Protein Sci., 29, 2020
|
|
2ALG
 
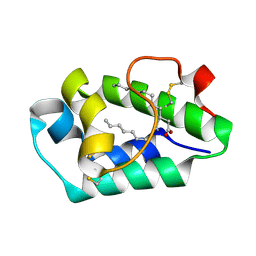 | | Crystal structure of peach Pru p3, the prototypic member of the family of plant non-specific lipid transfer protein pan-allergens | | 分子名称: | HEPTANE, HEXAETHYLENE GLYCOL, LAURIC ACID, ... | | 著者 | Pasquato, N, Berni, R, Folli, C, Folloni, S, Cianci, M, Pantano, S, Helliwell, J, Zanotti, G. | | 登録日 | 2005-08-05 | | 公開日 | 2005-11-29 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of Peach Pru p 3, the Prototypic Member of the Family of Plant Non-specific Lipid Transfer Protein Pan-allergens
J.Mol.Biol., 356, 2006
|
|
2ASK
 
 | | Structure of human Artemin | | 分子名称: | SULFATE ION, artemin | | 著者 | Silvian, L, Jin, P, Carmillo, P, Boriack-Sjodin, P.A, Pelletier, C, Rushe, M, Gong, B.J, Sah, D, Pepinsky, B, Rossomando, A. | | 登録日 | 2005-08-23 | | 公開日 | 2006-06-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Artemin crystal structure reveals insights into heparan sulfate binding.
Biochemistry, 45, 2006
|
|
1R1W
 
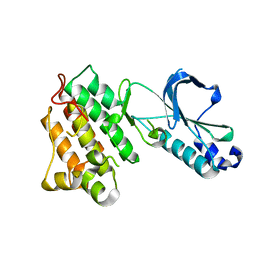 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HEPATOCYTE GROWTH FACTOR RECEPTOR C-MET | | 分子名称: | HEPATOCYTE GROWTH FACTOR RECEPTOR | | 著者 | Schiering, N, Knapp, S, Marconi, M, Flocco, M.M, Cui, J, Perego, R, Rusconi, L, Cristiani, C. | | 登録日 | 2003-09-25 | | 公開日 | 2003-10-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2AMQ
 
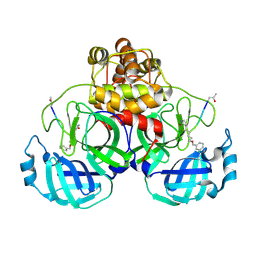 | | Crystal Structure Of SARS_CoV Mpro in Complex with an Inhibitor N3 | | 分子名称: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | 著者 | Yang, H, Xue, X, Yang, K, Zhao, Q, Bartlam, M, Rao, Z. | | 登録日 | 2005-08-10 | | 公開日 | 2005-09-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
1R26
 
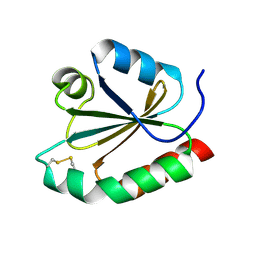 | | Crystal structure of thioredoxin from Trypanosoma brucei brucei | | 分子名称: | Thioredoxin | | 著者 | Friemann, R, Schmidt, H, Ramaswamy, S, Forstner, M, Krauth-Siegel, R.L, Eklund, H. | | 登録日 | 2003-09-26 | | 公開日 | 2003-12-09 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure of thioredoxin from Trypanosoma brucei brucei
FEBS Lett., 554, 2003
|
|
6WEN
 
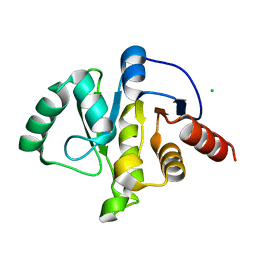 | | Crystal Structure of ADP ribose phosphatase of NSP3 from SARS-CoV-2 in the apo form | | 分子名称: | CHLORIDE ION, Non-structural protein 3 | | 著者 | Michalska, K, Stols, L, Jedrzejczak, R, Endres, M, Babnigg, G, Kim, Y, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-02 | | 公開日 | 2020-04-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal structures of SARS-CoV-2 ADP-ribose phosphatase: from the apo form to ligand complexes.
Iucrj, 7, 2020
|
|
