5GTB
 
 | |
3HVL
 
 | | Tethered PXR-LBD/SRC-1p complexed with SR-12813 | | 分子名称: | Pregnane X receptor, Linker, Steroid receptor coactivator 1, ... | | 著者 | Lesburg, C.A, Wang, W, Prosise, W.W, Chen, J, Taremi, S.S, Le, H.V, Madison, V, Cui, X, Thomas, A, Cheng, K.C. | | 登録日 | 2009-06-16 | | 公開日 | 2009-08-04 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Construction and characterization of a fully active PXR/SRC-1 tethered protein with increased stability
Protein Eng.Des.Sel., 21, 2008
|
|
3NOJ
 
 | | The structure of HMG/CHA aldolase from the protocatechuate degradation pathway of Pseudomonas putida | | 分子名称: | 4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetate decarboxylase, MAGNESIUM ION, PYRUVIC ACID, ... | | 著者 | Kimber, M.S, Wang, W, Mazurkewich, S, Seah, S.Y.K. | | 登録日 | 2010-06-25 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structural and Kinetic Characterization of 4-Hydroxy-4-methyl-2-oxoglutarate/4-Carboxy-4-hydroxy-2-oxoadipate Aldolase, a Protocatechuate Degradation Enzyme Evolutionarily Convergent with the HpaI and DmpG Pyruvate Aldolases.
J.Biol.Chem., 285, 2010
|
|
4ZMH
 
 | | Crystal structure of a five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T | | 分子名称: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Nocek, B, Cui, H, Wang, W, Savchenko, A. | | 登録日 | 2015-05-04 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Biochemical and Structural Characterization of a Five-domain GH115 alpha-Glucuronidase from the Marine Bacterium Saccharophagus degradans 2-40T.
J.Biol.Chem., 291, 2016
|
|
8P5Y
 
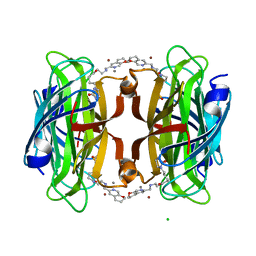 | | Artificial transfer hydrogenase with a Mn-12 cofactor and Streptavidin S112Y-K121M mutant | | 分子名称: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)ethyl]pentanamide, BROMIDE ION, CHLORIDE ION, ... | | 著者 | Lau, K, Wang, W, Pojer, F, Larabi, A. | | 登録日 | 2023-05-24 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8P5Z
 
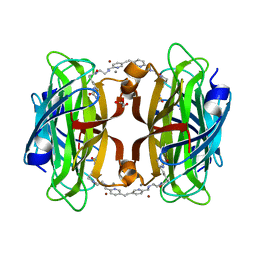 | | Artificial transfer hydrogenase with a Mn-5 cofactor and Streptavidin S112Y-K121M mutant | | 分子名称: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[2-[(5-methylpyridin-2-yl)methylamino]ethyl]pentanamide, BROMIDE ION, GLYCEROL, ... | | 著者 | Lau, K, Wang, W, Pojer, F, Larabi, A. | | 登録日 | 2023-05-24 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Manganese Transfer Hydrogenases Based on the Biotin-Streptavidin Technology.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7XUR
 
 | |
4I35
 
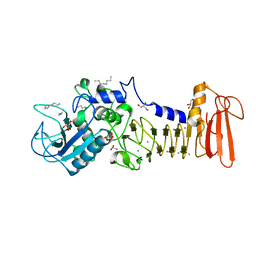 | | The crystal structure of serralysin | | 分子名称: | CALCIUM ION, GLYCEROL, HEXANE, ... | | 著者 | Zou, M, Ran, T, Xu, D, Wang, W. | | 登録日 | 2012-11-24 | | 公開日 | 2013-11-27 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.501 Å) | | 主引用文献 | The crystal structure of serralysin
To be Published
|
|
4O19
 
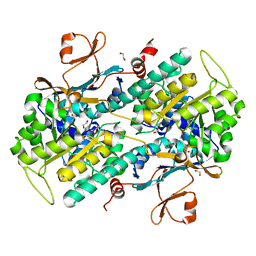 | | The crystal structure of a mutant NAMPT (G217V) | | 分子名称: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-15 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O13
 
 | | The crystal structure of NAMPT in complex with GNE-618 | | 分子名称: | 1,2-ETHANEDIOL, N-(4-{[3-(trifluoromethyl)phenyl]sulfonyl}benzyl)-2H-pyrazolo[3,4-b]pyridine-5-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-15 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O8P
 
 | | Crystal structure of SthAraf62A, a GH62 family alpha-L-arabinofuranosidase from Streptomyces thermoviolaceus, bound to xylotetraose | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Alpha-L-arabinofuranosidase, CALCIUM ION, ... | | 著者 | Stogios, P.J, Wang, W, Xu, X, Cui, H, Master, E, Savchenko, A. | | 登録日 | 2013-12-28 | | 公開日 | 2014-07-02 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.557 Å) | | 主引用文献 | Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 alpha-l-arabinofuranosidase from Streptomyces thermoviolaceus.
Appl.Environ.Microbiol., 80, 2014
|
|
4FXX
 
 | | Structure of SF1 coiled-coil domain | | 分子名称: | IMIDAZOLE, MALONATE ION, Splicing factor 1 | | 著者 | Gupta, A, Bauer, W.J, Wang, W, Kielkopf, C.L. | | 登録日 | 2012-07-03 | | 公開日 | 2013-01-16 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.4801 Å) | | 主引用文献 | Structure of Phosphorylated SF1 Bound to U2AF(65) in an Essential Splicing Factor Complex.
Structure, 21, 2013
|
|
4O1C
 
 | | The crystal structures of a mutant NAMPT H191R | | 分子名称: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-15 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.092 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O14
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | 分子名称: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-15 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.871 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O17
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | 分子名称: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-15 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O28
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | 分子名称: | 1,2-ETHANEDIOL, N-{4-[(3,5-difluorophenyl)sulfonyl]benzyl}imidazo[1,2-a]pyridine-6-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-17 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O1A
 
 | | The crystal structure of the mutant NAMPT G217R | | 分子名称: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-15 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O18
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | 分子名称: | 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, PHOSPHATE ION | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-15 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O1B
 
 | | The crystal structure of a mutant NAMPT (G217R) in complex with an inhibitor APO866 | | 分子名称: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-15 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O1D
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | 分子名称: | (2E)-N-{4-[1-(benzenecarbonyl)piperidin-4-yl]butyl}-3-(pyridin-3-yl)prop-2-enamide, 1,2-ETHANEDIOL, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-15 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.705 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O15
 
 | | The crystal structure of a mutant NAMPT (S165F) in complex with GNE-618 | | 分子名称: | 1,2-ETHANEDIOL, N-(4-{[3-(trifluoromethyl)phenyl]sulfonyl}benzyl)-2H-pyrazolo[3,4-b]pyridine-5-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-15 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O16
 
 | | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors | | 分子名称: | 1,2-ETHANEDIOL, 6-({4-[(3,5-difluorophenyl)sulfonyl]benzyl}carbamoyl)-1-(5-O-phosphono-beta-D-ribofuranosyl)imidazo[1,2-a]pyridin-1-ium, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Oh, A, Coons, M, Brillantes, B, Wang, W. | | 登録日 | 2013-12-15 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.783 Å) | | 主引用文献 | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
4O8N
 
 | | Crystal structure of SthAraf62A, a GH62 family alpha-L-arabinofuranosidase from Streptomyces thermoviolaceus, in the apoprotein form | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, CALCIUM ION, ... | | 著者 | Stogios, P.J, Wang, W, Xu, X, Cui, H, Master, E, Savchenko, A. | | 登録日 | 2013-12-28 | | 公開日 | 2014-07-02 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.6476 Å) | | 主引用文献 | Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 alpha-l-arabinofuranosidase from Streptomyces thermoviolaceus.
Appl.Environ.Microbiol., 80, 2014
|
|
4O8O
 
 | | Crystal structure of SthAraf62A, a GH62 family alpha-L-arabinofuranosidase from Streptomyces thermoviolaceus, bound to alpha-L-arabinose | | 分子名称: | Alpha-L-arabinofuranosidase, CALCIUM ION, alpha-L-arabinofuranose | | 著者 | Stogios, P.J, Wang, W, Xu, X, Cui, H, Master, E, Savchenko, A. | | 登録日 | 2013-12-28 | | 公開日 | 2014-07-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 alpha-l-arabinofuranosidase from Streptomyces thermoviolaceus.
Appl.Environ.Microbiol., 80, 2014
|
|
5WVM
 
 | | Crystal structure of baeS cocrystallized with 2 mM indole | | 分子名称: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | 著者 | Wang, W, Zhang, Y, Rang, T, Xu, D. | | 登録日 | 2016-12-26 | | 公開日 | 2018-01-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
