8SAI
 
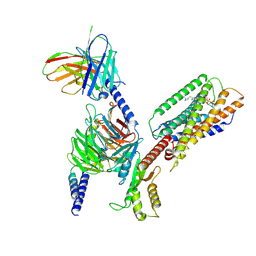 | | Cryo-EM structure of GPR34-Gi complex | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Yong, X.H, Zhao, C, Yan, W, Shao, Z.H. | | 登録日 | 2023-04-01 | | 公開日 | 2023-10-04 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3IN4
 
 | | Bace1 with Compound 38 | | 分子名称: | (5S)-2-amino-5-(2,6-diethylpyridin-4-yl)-3-methyl-5-(3-pyrimidin-5-ylphenyl)-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | 著者 | Olland, A.M. | | 登録日 | 2009-08-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Di-substituted pyridinyl aminohydantoins as potent and highly selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem., 18, 2010
|
|
3IN3
 
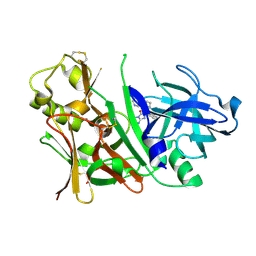 | | Bace1 with Compound 30 | | 分子名称: | (5S)-2-amino-3-methyl-5-pyridin-4-yl-5-(3-pyridin-3-ylphenyl)-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | 著者 | Olland, A.M. | | 登録日 | 2009-08-11 | | 公開日 | 2010-01-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Di-substituted pyridinyl aminohydantoins as potent and highly selective human beta-secretase (BACE1) inhibitors.
Bioorg.Med.Chem., 18, 2010
|
|
8DTF
 
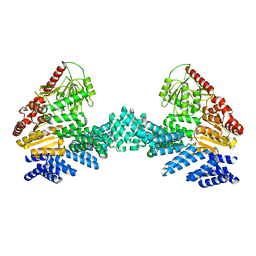 | | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs | | 分子名称: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | 著者 | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | 登録日 | 2022-07-25 | | 公開日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTH
 
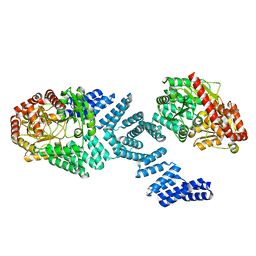 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 2 | | 分子名称: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | 著者 | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | 登録日 | 2022-07-25 | | 公開日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTI
 
 | | Cryo-EM structure of Arabidopsis SPY in complex with GDP-fucose | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | 著者 | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | 登録日 | 2022-07-25 | | 公開日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTG
 
 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 1 | | 分子名称: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | 著者 | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | 登録日 | 2022-07-25 | | 公開日 | 2023-03-08 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
5W5D
 
 | |
5W5C
 
 | |
6A67
 
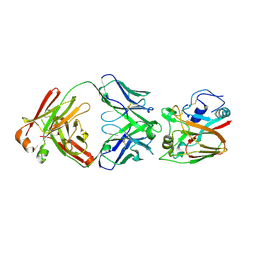 | | Crystal structure of influenza A virus H5 hemagglutinin globular head in complex with the Fab of antibody FLD21.140 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLD21.140 Heavy Chain, FLD21.140 Light Chain, ... | | 著者 | Wang, P, Zuo, Y, Sun, J, Zhang, L, Wang, X. | | 登録日 | 2018-06-26 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Complementary recognition of the receptor-binding site of highly pathogenic H5N1 influenza viruses by two human neutralizing antibodies.
J. Biol. Chem., 293, 2018
|
|
8SIT
 
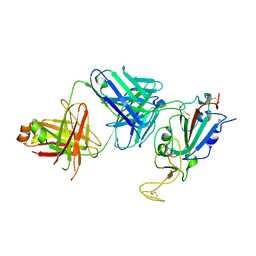 | |
8SIR
 
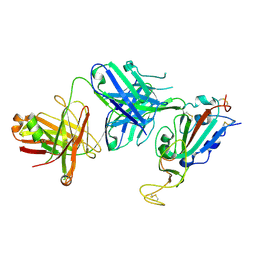 | |
8SIQ
 
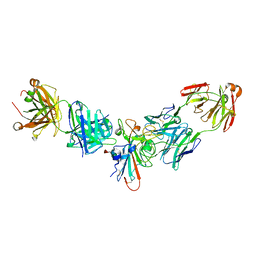 | |
8SDF
 
 | |
8SDG
 
 | |
8SIS
 
 | |
3NK7
 
 | | Structure of the Nosiheptide-resistance methyltransferase S-adenosyl-L-methionine Complex | | 分子名称: | 23S rRNA methyltransferase, S-ADENOSYLMETHIONINE | | 著者 | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | 登録日 | 2010-06-18 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
3NK6
 
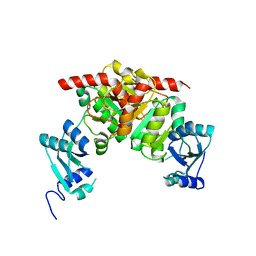 | | Structure of the Nosiheptide-resistance methyltransferase | | 分子名称: | 23S rRNA methyltransferase | | 著者 | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | 登録日 | 2010-06-18 | | 公開日 | 2010-07-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
3OOZ
 
 | | Bace1 in complex with the aminohydantoin Compound 102 | | 分子名称: | (5R)-2-amino-5-[4-(difluoromethoxy)phenyl]-5-[4-fluoro-3-(5-fluoropent-1-yn-1-yl)phenyl]-3-methyl-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | 著者 | Olland, A.M. | | 登録日 | 2010-08-31 | | 公開日 | 2011-08-31 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Design and Synthesis of Aminohydantoins as Potent and Selective Human beta-Secretase (BACE1) Inhibitors with Enhanced Brain Permeability
Bioorg.Med.Chem.Lett., 20, 2010
|
|
8IYX
 
 | | Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 | | 分子名称: | 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid, Probable G-protein coupled receptor 34,YL-365 | | 著者 | Jia, G.W, Wang, X, Zhang, C.B, Dong, H.H, Su, Z.M. | | 登録日 | 2023-04-06 | | 公開日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.34 Å) | | 主引用文献 | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5ZXV
 
 | |
7RU5
 
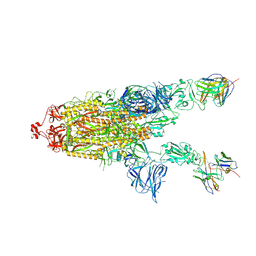 | | CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.30 Fab heavy chain Fv, ... | | 著者 | Ozorowski, G, Turner, H.L, Ward, A.B. | | 登録日 | 2021-08-16 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU2
 
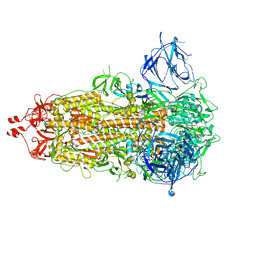 | | SARS-CoV-2-6P-Mut7 S protein (asymmetric) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Ozorowski, G, Turner, H.L, Ward, A.B. | | 登録日 | 2021-08-16 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU8
 
 | | CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.30 Fab Kappa chain Fv, ... | | 著者 | Ozorowski, G, Turner, H.L, Ward, A.B. | | 登録日 | 2021-08-16 | | 公開日 | 2022-08-24 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU3
 
 | | CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Ozorowski, G, Turner, H.L, Ward, A.B. | | 登録日 | 2021-08-16 | | 公開日 | 2022-08-24 | | 最終更新日 | 2022-09-14 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
