5TUH
 
 | |
1YHH
 
 | |
5TUG
 
 | |
3SR2
 
 | | Crystal Structure of Human XLF-XRCC4 Complex | | 分子名称: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | 著者 | Hammel, M, Classen, S, Tainer, J.A. | | 登録日 | 2011-07-06 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (3.9708 Å) | | 主引用文献 | XRCC4 Protein Interactions with XRCC4-like Factor (XLF) Create an Extended Grooved Scaffold for DNA Ligation and Double Strand Break Repair.
J.Biol.Chem., 286, 2011
|
|
3SOD
 
 | | CHANGES IN CRYSTALLOGRAPHIC STRUCTURE AND THERMOSTABILITY OF A CU,ZN SUPEROXIDE DISMUTASE MUTANT RESULTING FROM THE REMOVAL OF BURIED CYSTEINE | | 分子名称: | COPPER (II) ION, COPPER,ZINC SUPEROXIDE DISMUTASE, ZINC ION | | 著者 | Mcree, D.E, Redford, S.M, Getzoff, E.D, Lepock, J.R, Hallewell, R.A, Tainer, J.A. | | 登録日 | 1990-06-26 | | 公開日 | 1993-04-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Changes in crystallographic structure and thermostability of a Cu,Zn superoxide dismutase mutant resulting from the removal of a buried cysteine.
J.Biol.Chem., 265, 1990
|
|
3SOJ
 
 | | Francisella tularensis pilin PilE | | 分子名称: | PilE, SULFATE ION | | 著者 | Wood, T, Arvai, A.S, Shin, D.S, Hartung, S, Kolappan, S, Craig, L, Tainer, J.A. | | 登録日 | 2011-06-30 | | 公開日 | 2011-11-02 | | 最終更新日 | 2014-05-14 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Ultrahigh Resolution and Full-length Pilin Structures with Insights for Filament Assembly, Pathogenic Functions, and Vaccine Potential.
J.Biol.Chem., 286, 2011
|
|
3UEL
 
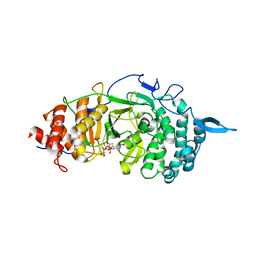 | | Crystal structure of the catalytic domain of rat poly (ADP-ribose) glycohydrolase bound to ADP-HPD | | 分子名称: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, Poly(ADP-ribose) glycohydrolase | | 著者 | Kim, I.K, Kiefer, J.R, Stegemann, R.A, Classen, S, Tainer, J.A, Ellenberger, T. | | 登録日 | 2011-10-30 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UMV
 
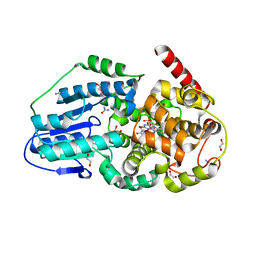 | | Eukaryotic Class II CPD photolyase structure reveals a basis for improved UV-tolerance in plants | | 分子名称: | 1,2-ETHANEDIOL, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Arvai, A.S, Hitomi, K, Getzoff, E.D, Tainer, J.A. | | 登録日 | 2011-11-14 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.705 Å) | | 主引用文献 | Eukaryotic Class II Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals Basis for Improved Ultraviolet Tolerance in Plants.
J.Biol.Chem., 287, 2012
|
|
3SOK
 
 | | Dichelobacter nodosus pilin FimA | | 分子名称: | Fimbrial protein | | 著者 | Arvai, A.S, Craig, L, Hartung, S, Wood, T, Kolappan, S, Shin, D.S, Tainer, J.A. | | 登録日 | 2011-06-30 | | 公開日 | 2011-11-02 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Ultrahigh Resolution and Full-length Pilin Structures with Insights for Filament Assembly, Pathogenic Functions, and Vaccine Potential.
J.Biol.Chem., 286, 2011
|
|
3UEK
 
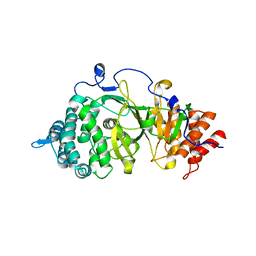 | | Crystal structure of the catalytic domain of rat poly (ADP-ribose) glycohydrolase | | 分子名称: | Poly(ADP-ribose) glycohydrolase | | 著者 | Kim, I.K, Kiefer, J.R, Stegemann, R.A, Classen, S, Tainer, J.A, Ellenberger, T. | | 登録日 | 2011-10-30 | | 公開日 | 2012-05-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure of mammalian poly(ADP-ribose) glycohydrolase reveals a flexible tyrosine clasp as a substrate-binding element.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VHJ
 
 | |
1UGI
 
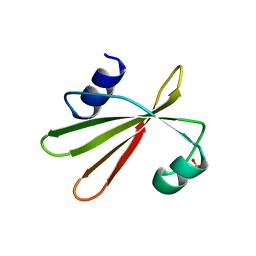 | | URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | 分子名称: | IMIDAZOLE, SULFATE ION, URACIL-DNA GLYCOSYLASE INHIBITOR | | 著者 | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | 登録日 | 1998-11-04 | | 公開日 | 1999-03-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
1UUG
 
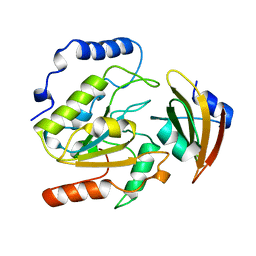 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI | | 分子名称: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | 著者 | Mol, C.D, Arvai, A.S, Putnam, C.D, Tainer, J.A. | | 登録日 | 1998-10-31 | | 公開日 | 1999-03-25 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
1WEG
 
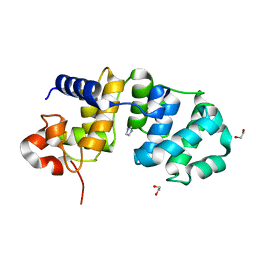 | | Catalytic Domain Of Muty From Escherichia Coli K142A Mutant | | 分子名称: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, IMIDAZOLE, ... | | 著者 | Hitomi, K, Arvai, A.S, Tainer, J.A. | | 登録日 | 2004-05-25 | | 公開日 | 2004-09-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
3HVC
 
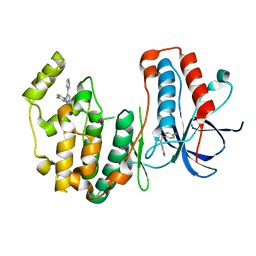 | | Crystal structure of human p38alpha MAP kinase | | 分子名称: | 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, Mitogen-activated protein kinase 14 | | 著者 | Perry, J.J, Tainer, J.A. | | 登録日 | 2009-06-15 | | 公開日 | 2009-06-30 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | p38alpha MAP kinase C-terminal domain binding pocket characterized by crystallographic and computational analyses.
J.Mol.Biol., 391, 2009
|
|
3FY4
 
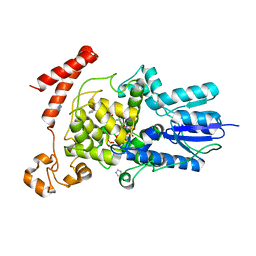 | | (6-4) Photolyase Crystal Structure | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-4 photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Hitomi, K, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2009-01-21 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1WEF
 
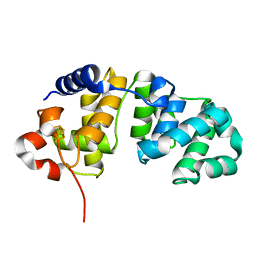 | |
1WEI
 
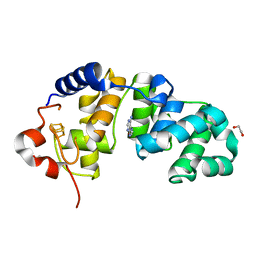 | | Catalytic Domain Of Muty From Escherichia Coli K20A Mutant Complexed To Adenine | | 分子名称: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, ADENINE, ... | | 著者 | Hitomi, K, Arvai, A.S, Tainer, J.A. | | 登録日 | 2004-05-25 | | 公開日 | 2004-09-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
3HUE
 
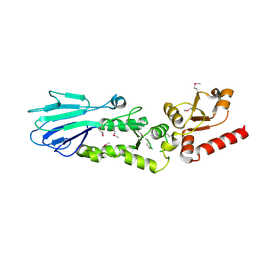 | |
3HUF
 
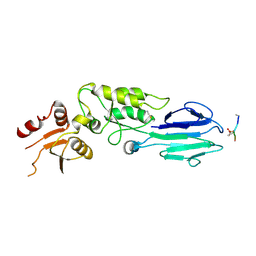 | | Structure of the S. pombe Nbs1-Ctp1 complex | | 分子名称: | DNA repair and telomere maintenance protein nbs1, Double-strand break repair protein ctp1, THIOCYANATE ION | | 著者 | Williams, R.S, Guenther, G, Tainer, J.A. | | 登録日 | 2009-06-13 | | 公開日 | 2009-10-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair.
Cell(Cambridge,Mass.), 139, 2009
|
|
3HRV
 
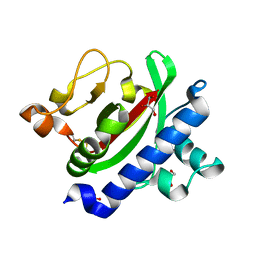 | | Crystal structure of TcpA, a Type IV pilin from Vibrio cholerae El Tor biotype | | 分子名称: | GLYCEROL, SULFATE ION, Toxin coregulated pilin | | 著者 | Craig, L, Arvai, A.S, Tainer, J.A. | | 登録日 | 2009-06-09 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Vibrio cholerae El Tor TcpA crystal structure and mechanism for pilus-mediated microcolony formation.
Mol.Microbiol., 77, 2010
|
|
1XDC
 
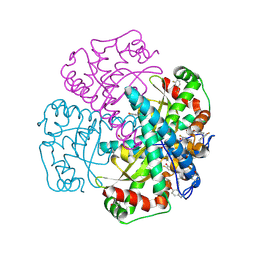 | | Hydrogen Bonding in Human Manganese Superoxide Dismutase containing 3-Fluorotyrosine | | 分子名称: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | 著者 | Ayala, I, Perry, J.J, Szczepanski, J, Cabelli, D.E, Tainer, J.A, Vala, M.T, Nick, H.S, Silverman, D.N. | | 登録日 | 2004-09-05 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Hydrogen bonding in human manganese superoxide dismutase containing 3-fluorotyrosine
Biophys.J., 89, 2005
|
|
3GVA
 
 | |
3GX4
 
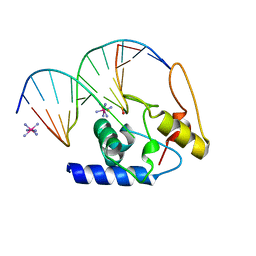 | | Crystal Structure Analysis of S. Pombe ATL in complex with DNA | | 分子名称: | Alkyltransferase-like protein 1, COBALT HEXAMMINE(III), DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | 著者 | Tubbs, J.L, Arvai, A.S, Tainer, J.A. | | 登録日 | 2009-04-01 | | 公開日 | 2009-06-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
3GYH
 
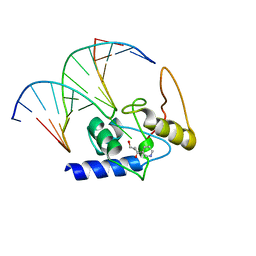 | | Crystal Structure Analysis of S. Pombe ATL in complex with damaged DNA containing POB | | 分子名称: | 1-PYRIDIN-3-YLBUTAN-1-ONE, Alkyltransferase-like protein 1, DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | 著者 | Tubbs, J.L, Arvai, A.S, Tainer, J.A, Shin, D.S. | | 登録日 | 2009-04-03 | | 公開日 | 2009-06-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
