1DYB
 
 | |
1DYE
 
 | |
1DYA
 
 | |
1DYG
 
 | |
1DP0
 
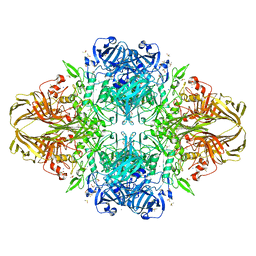 | | E. COLI BETA-GALACTOSIDASE AT 1.7 ANGSTROM | | 分子名称: | BETA-GALACTOSIDASE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | 著者 | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | 登録日 | 1999-12-22 | | 公開日 | 2001-02-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
1D2Y
 
 | |
3CDQ
 
 | |
2OE9
 
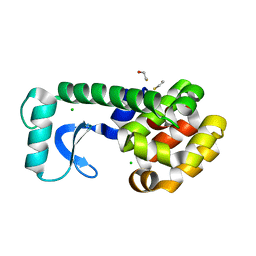 | | High-pressure structure of pseudo-WT T4 Lysozyme | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | 著者 | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | 登録日 | 2006-12-28 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
1XPX
 
 | |
3F8V
 
 | |
3FA0
 
 | |
2OE7
 
 | | High-Pressure T4 Lysozyme | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | 著者 | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | 登録日 | 2006-12-28 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
2OEA
 
 | | High-pressure structure of pseudo-WT T4 Lysozyme | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | 著者 | Collins, M.D, Quillin, M.L, Matthews, B.W, Gruner, S.M. | | 登録日 | 2006-12-28 | | 公開日 | 2007-01-30 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural Rigidity of a Large Cavity-containing Protein Revealed by High-pressure Crystallography.
J.Mol.Biol., 367, 2007
|
|
3FAD
 
 | |
1Y1N
 
 | | Identification of SH3 motif in M. Tuberculosis methionine aminopeptidase suggests a mode of interaction with the ribosome | | 分子名称: | Methionine aminopeptidase 1B, POTASSIUM ION | | 著者 | Addlagatta, A, Quillin, M.L, Omotoso, O, Liu, J.O, Matthews, B.W. | | 登録日 | 2004-11-18 | | 公開日 | 2005-05-24 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Identification of an SH3-Binding Motif in a New Class of Methionine Aminopeptidases from Mycobacterium tuberculosis Suggests a Mode of Interaction with the Ribosome
Biochemistry, 44, 2005
|
|
3FI5
 
 | |
152L
 
 | |
3F9L
 
 | |
3BBZ
 
 | | Structure of the nucleocapsid-binding domain from the mumps virus phosphoprotein | | 分子名称: | BROMIDE ION, FORMIC ACID, P protein | | 著者 | Kingston, R.L, Gay, L.S, Baase, W.S, Matthews, B.W. | | 登録日 | 2007-11-11 | | 公開日 | 2008-05-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the nucleocapsid-binding domain from the mumps virus polymerase; an example of protein folding induced by crystallization
J.Mol.Biol., 379, 2008
|
|
3B2X
 
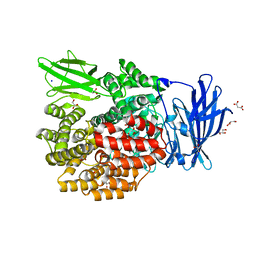 | | Crystal Structure of E. coli Aminopeptidase N in complex with Lysine | | 分子名称: | Aminopeptidase N, GLYCEROL, LYSINE, ... | | 著者 | Addlagatta, A, Gay, L, Matthews, B.W. | | 登録日 | 2007-10-19 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis for the unusual specificity of Escherichia coli aminopeptidase N.
Biochemistry, 47, 2008
|
|
1CU0
 
 | | T4 LYSOZYME MUTANT I78M | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | 著者 | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | 登録日 | 1999-08-20 | | 公開日 | 1999-11-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CV4
 
 | | T4 LYSOZYME MUTANT L118M | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | 著者 | Gassner, N.C, Baase, W.A, Lindstrom, J, Lu, J, Matthews, B.W. | | 登録日 | 1999-08-22 | | 公開日 | 1999-11-10 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
3GUP
 
 | |
3GUJ
 
 | |
3GUO
 
 | |
