7OQM
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.05 Angstroms resolution, 20 minutes soaking | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | 登録日 | 2021-06-03 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQN
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.10 Angstroms resolution, 30 minutes soaking | | 分子名称: | GLYCEROL, SULFATE ION, URIDINE-5'-MONOPHOSPHATE, ... | | 著者 | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | 登録日 | 2021-06-03 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQF
 
 | | Human OMPD-domain of UMPS in complex with OMP at 1.05 Angstrom resolution, 5 minutes soaking | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | 登録日 | 2021-06-03 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQI
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.15 Angstrom resolution, 10 minutes soaking | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | 登録日 | 2021-06-03 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OQK
 
 | | Human OMPD-domain of UMPS in complex with substrate OMP at 1.10 Angstroms resolution, 15 minutes soaking | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | 登録日 | 2021-06-03 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7OUZ
 
 | |
7OTU
 
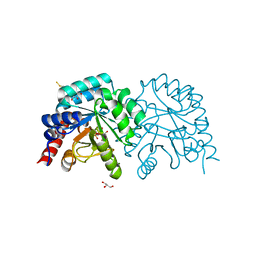 | | Human OMPD-domain of UMPS in complex with 6-hydroxy-UMP at 0.95 Angstroms resolution, crystal 2 | | 分子名称: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Isoform 2 of Uridine 5'-monophosphate synthase | | 著者 | Rindfleisch, S, Rabe von Pappenheim, F, Tittmann, K. | | 登録日 | 2021-06-10 | | 公開日 | 2022-04-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.95 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
1MI1
 
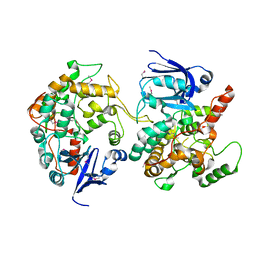 | | Crystal Structure of the PH-BEACH Domain of Human Neurobeachin | | 分子名称: | Neurobeachin | | 著者 | Jogl, G, Shen, Y, Gebauer, D, Li, J, Wiegmann, K, Kashkar, H, Kroenke, M, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2002-08-21 | | 公開日 | 2002-09-27 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain.
EMBO J., 21, 2002
|
|
5HHT
 
 | |
1HPY
 
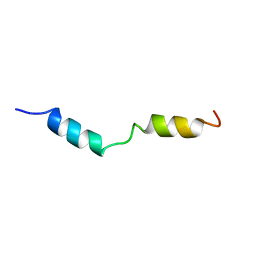 | | THE SOLUTION STRUCTURE OF HUMAN PARATHYROID HORMONE FRAGMENT 1-34 IN 20% TRIFLUORETHANOL, NMR, 10 STRUCTURES | | 分子名称: | PARATHYROID HORMONE | | 著者 | Marx, U.C, Roesch, P, Adermann, K, Bayer, P, Forssmann, W.-G. | | 登録日 | 1998-09-30 | | 公開日 | 2000-01-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of human parathyroid hormone fragments hPTH(1-34) and hPTH(1-39) and bovine parathyroid hormone fragment bPTH(1-37).
Biochem.Biophys.Res.Commun., 267, 2000
|
|
7BBX
 
 | | Neisseria gonorrhoeae transaldolase, variant K8A | | 分子名称: | 1,2-ETHANEDIOL, CITRIC ACID, Transaldolase | | 著者 | Rabe von Pappenheim, F, Wensien, M, Funk, L.M, Tittmann, K. | | 登録日 | 2020-12-18 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | A lysine-cysteine redox switch with an NOS bridge regulates enzyme function.
Nature, 593, 2021
|
|
7BBW
 
 | |
2HCC
 
 | | SOLUTION STRUCTURE OF THE HUMAN CHEMOKINE HCC-2, NMR, 30 STRUCTURES | | 分子名称: | HUMAN CHEMOKINE HCC-2 | | 著者 | Sticht, H, Escher, S.E, Schweimer, K, Forssmann, W.G, Roesch, P, Adermann, K. | | 登録日 | 1998-07-03 | | 公開日 | 1999-07-13 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the human CC chemokine 2: A monomeric representative of the CC chemokine subtype.
Biochemistry, 38, 1999
|
|
3FZN
 
 | | Intermediate analogue in benzoylformate decarboxylase | | 分子名称: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, CHLORIDE ION, ... | | 著者 | Bruning, M, Berheide, M, Meyer, D, Golbik, R, Bartunik, H, Liese, A, Tittmann, K. | | 登録日 | 2009-01-26 | | 公開日 | 2009-05-05 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Structural and kinetic studies on native intermediates and an intermediate analogue in benzoylformate decarboxylase reveal a least motion mechanism with an unprecedented short-lived predecarboxylation intermediate.
Biochemistry, 48, 2009
|
|
8CMR
 
 | | Linear specific OTU-type DUB SnOTU from the pathogen S. negenvensis in complex with linear di-ubiquitin | | 分子名称: | OTU domain-containing protein, Polyubiquitin-B | | 著者 | Uthoff, M, Hermanns, T, Boll, V, Hofmann, K, Baumann, U. | | 登録日 | 2023-02-21 | | 公開日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Functional and structural diversity in deubiquitinases of the Chlamydia-like bacterium Simkania negevensis.
Nat Commun, 14, 2023
|
|
3IO1
 
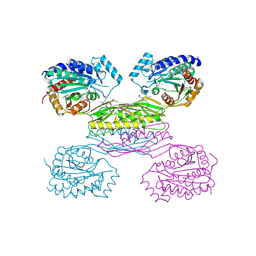 | | Crystal Structure of Aminobenzoyl-glutamate utilization protein from Klebsiella pneumoniae | | 分子名称: | Aminobenzoyl-glutamate utilization protein, SODIUM ION, YTTRIUM (III) ION | | 著者 | Kumaran, D, Baumann, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2009-08-13 | | 公開日 | 2009-08-25 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of Aminobenzoyl-glutamate utilization protein from Klebsiella pneumoniae
To be Published
|
|
6RJC
 
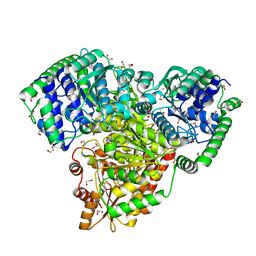 | |
6HAF
 
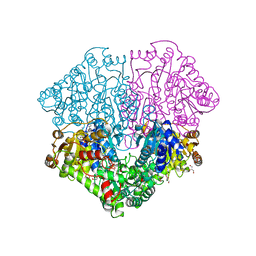 | | Pyruvate oxidase variant E59Q from L. plantarum in complex with phosphate | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Funk, L.M, Sautner, V, Tittmann, K. | | 登録日 | 2018-08-07 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Low-barrier hydrogen bonds in enzyme cooperativity.
Nature, 573, 2019
|
|
6HA3
 
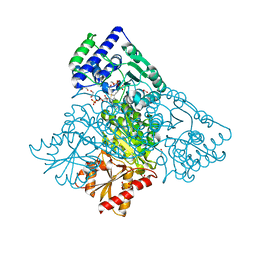 | | Human transketolase variant E160Q in covalent complex with donor ketose D-fructose-6-phosphate | | 分子名称: | 1,2-ETHANEDIOL, 2-C-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium-2-yl}-6-O-phosphono-D-glucitol, CALCIUM ION, ... | | 著者 | Dai, S, Sautner, V, Tittmann, K. | | 登録日 | 2018-08-07 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Low-barrier hydrogen bonds in enzyme cooperativity.
Nature, 573, 2019
|
|
6H55
 
 | | core of the human pyruvate dehydrogenase (E2) | | 分子名称: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | 著者 | Haselbach, D, Prajapati, S, Tittmann, K, Stark, H. | | 登録日 | 2018-07-23 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Structural and Functional Analyses of the Human PDH Complex Suggest a "Division-of-Labor" Mechanism by Local E1 and E3 Clusters.
Structure, 27, 2019
|
|
6H60
 
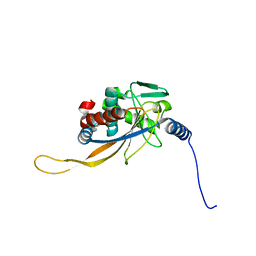 | | pseudo-atomic structural model of the E3BP component of the human pyruvate dehydrogenase multienzyme complex | | 分子名称: | Pyruvate dehydrogenase protein X component, mitochondrial | | 著者 | Haselbach, D, Prajapati, S, Tittmann, K, Stark, H. | | 登録日 | 2018-07-25 | | 公開日 | 2019-06-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Structural and Functional Analyses of the Human PDH Complex Suggest a "Division-of-Labor" Mechanism by Local E1 and E3 Clusters.
Structure, 27, 2019
|
|
6RJB
 
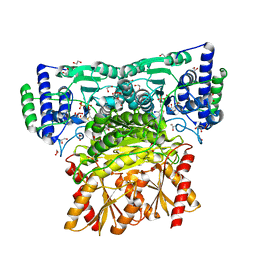 | |
6HAD
 
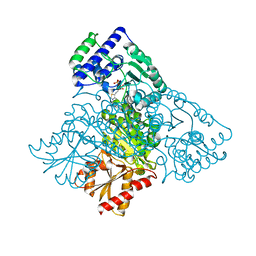 | | Human transketolase variant E160Q | | 分子名称: | CALCIUM ION, MAGNESIUM ION, SODIUM ION, ... | | 著者 | Dai, S, Sautner, V, Tittmann, K. | | 登録日 | 2018-08-07 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Low-barrier hydrogen bonds in enzyme cooperativity.
Nature, 573, 2019
|
|
1S2H
 
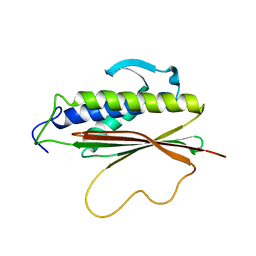 | | The Mad2 spindle checkpoint protein possesses two distinct natively folded states | | 分子名称: | Mitotic spindle assembly checkpoint protein MAD2A | | 著者 | Luo, X, Tang, Z, Xia, G, Wassmann, K, Matsumoto, T, Rizo, J, Yu, H. | | 登録日 | 2004-01-08 | | 公開日 | 2004-03-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Mad2 spindle checkpoint protein has two distinct natively folded states.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2QC7
 
 | | Crystal structure of the protein-disulfide isomerase related chaperone ERp29 | | 分子名称: | Endoplasmic reticulum protein ERp29 | | 著者 | Barak, N.N, Sevvana, M, Neumann, P, Malesevic, M, Naumann, K, Fischer, G, Sheldrick, G.M, Stubbs, M.T, Ferrari, D.M. | | 登録日 | 2007-06-19 | | 公開日 | 2008-06-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure and functional analysis of the protein disulfide isomerase-related protein ERp29.
J.Mol.Biol., 385, 2009
|
|
