7JH2
 
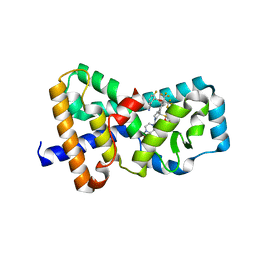 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C IN COMPLEX WITH A POTENT, SELECTIVE AND ORALLY BIOAVAILABLE ROR-GAMMA-T INVERSE AGONIST | | 分子名称: | 2-({[2-(4-{(3R)-1-(4-acetylpiperazine-1-carbonyl)-3-[(4-fluorophenyl)sulfonyl]pyrrolidin-3-yl}phenyl)-1,1,1,3,3,3-hexafluoropropan-2-yl]oxy}methyl)-3-fluorobenzonitrile, Nuclear receptor ROR-gamma, SULFATE ION | | 著者 | Sack, J.S. | | 登録日 | 2020-07-20 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.367 Å) | | 主引用文献 | Discovery of 2,6-difluorobenzyl ether series of phenyl ((R)-3-phenylpyrrolidin-3-yl)sulfones as surprisingly potent, selective and orally bioavailable ROR gamma t inverse agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7ZOI
 
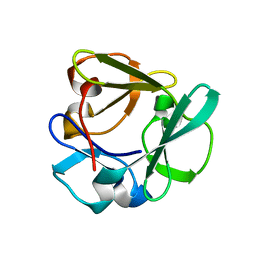 | | Carbohydrate binding domain CBM92-A from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 | | 分子名称: | Glycoside hydrolase family 18 | | 著者 | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | 登録日 | 2022-04-25 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOO
 
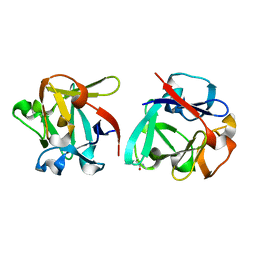 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with gentiobiose | | 分子名称: | Glycoside hydrolase family 18, beta-D-glucopyranose | | 著者 | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | 登録日 | 2022-04-26 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZON
 
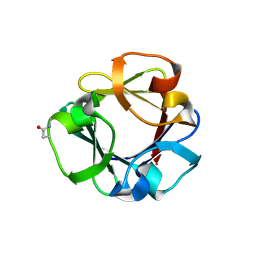 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with glucose | | 分子名称: | Glycoside hydrolase family 18, PENTAETHYLENE GLYCOL, beta-D-glucopyranose | | 著者 | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | 登録日 | 2022-04-26 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOH
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 | | 分子名称: | Glycoside hydrolase family 18 | | 著者 | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | 登録日 | 2022-04-25 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.56 Å) | | 主引用文献 | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
7ZOP
 
 | | Carbohydrate binding domain CBM92-B from a multi-catalytic glucanase-chitinase from Chitinophaga pinensis DSM 2588 in complex with sophorose. | | 分子名称: | Glycoside hydrolase family 18, beta-D-glucopyranose | | 著者 | Mazurkewich, S, McKee, L.S, Lu, Z, Branden, G, Larsbrink, J. | | 登録日 | 2022-04-26 | | 公開日 | 2023-05-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Structural and biochemical analysis of family 92 carbohydrate-binding modules uncovers multivalent binding to beta-glucans.
Nat Commun, 15, 2024
|
|
8HY0
 
 | |
8HXZ
 
 | |
8HXX
 
 | |
8HXY
 
 | |
8JLV
 
 | |
8JNX
 
 | |
8JHO
 
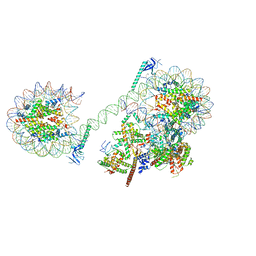 | |
2TRS
 
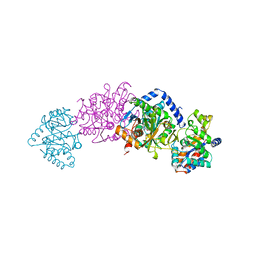 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | 分子名称: | INDOLE-3-PROPANOL PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | 著者 | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | 登録日 | 1997-01-07 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
2TSY
 
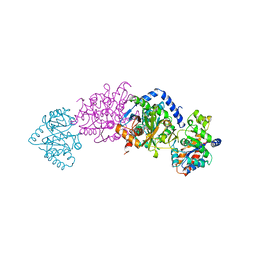 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | 分子名称: | SN-GLYCEROL-3-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE, ... | | 著者 | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | 登録日 | 1997-01-03 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
2TYS
 
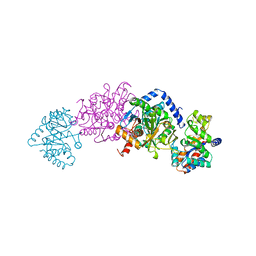 | | CRYSTAL STRUCTURES OF MUTANT (BETAK87T) TRYPTOPHAN SYNTHASE ALPHA2 BETA2 COMPLEX WITH LIGANDS BOUND TO THE ACTIVE SITES OF THE ALPHA AND BETA SUBUNITS REVEAL LIGAND-INDUCED CONFORMATIONAL CHANGES | | 分子名称: | SODIUM ION, TRYPTOPHAN SYNTHASE, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-L-TRYPTOPHANE | | 著者 | Rhee, S, Parris, K.D, Hyde, C.C, Ahmed, S.A, Miles, E.W, Davies, D.R. | | 登録日 | 1997-01-08 | | 公開日 | 1997-04-01 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes.
Biochemistry, 36, 1997
|
|
8H66
 
 | |
8H6D
 
 | |
8H6C
 
 | |
8H65
 
 | |
7WMK
 
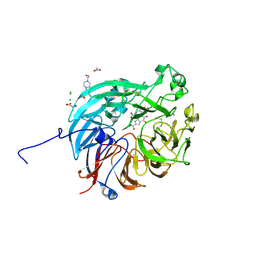 | | PQQ-dependent alcohol dehydrogenase complexed with PQQ | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | 著者 | Chen, M, Yang, H, Lv, F. | | 登録日 | 2022-01-15 | | 公開日 | 2022-09-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Structure-Function Analysis of a Quinone-Dependent Dehydrogenase Capable of Deoxynivalenol Detoxification.
J.Agric.Food Chem., 70, 2022
|
|
7WMD
 
 | |
7WLP
 
 | |
7W7P
 
 | |
7YHK
 
 | | Cryo-EM structure of the HA trimer of A/Beijing/262/1995(H1N1) in complex with neutralizing antibody 12H5 | | 分子名称: | 12H5 heavy chain, 12H5 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zheng, Q, Li, S, Li, T, Xue, W, Sun, H. | | 登録日 | 2022-07-13 | | 公開日 | 2022-08-17 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Identification of a cross-neutralizing antibody that targets the receptor binding site of H1N1 and H5N1 influenza viruses.
Nat Commun, 13, 2022
|
|
