3JYP
 
 | | Quinate dehydrogenase from Corynebacterium glutamicum in complex with quinate and NADH | | 分子名称: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Quinate/shikimate dehydrogenase | | 著者 | Hoeppner, A, Schomburg, D, Niefind, K. | | 登録日 | 2009-09-22 | | 公開日 | 2010-10-27 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Enzyme-substrate complexes of the quinate/shikimate dehydrogenase from Corynebacterium glutamicum enable new insights in substrate and cofactor binding, specificity, and discrimination.
Biol.Chem., 394, 2013
|
|
8B0G
 
 | | 2C9, C5b9-CD59 structure | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 glycoprotein, Complement C5, ... | | 著者 | Couves, E.C, Gardner, S, Bubeck, D. | | 登録日 | 2022-09-07 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-03-01 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
6TGD
 
 | | Crystal structure of NDM-1 in complex with triazole-based inhibitor OP31 | | 分子名称: | 4-[[(2~{S})-oxolan-2-yl]methyl]-3-pyridin-3-yl-1~{H}-1,2,4-triazole-5-thione, CALCIUM ION, Metallo-beta-lactamase NDM-1, ... | | 著者 | Maso, L, Spirakis, F, Santucci, M, Simon, C, Docquier, J.D, Cruciani, G, Costi, M.P, Tondi, D, Cendron, L. | | 登録日 | 2019-11-15 | | 公開日 | 2020-08-26 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Virtual screening identifies broad-spectrum beta-lactamase inhibitors with activity on clinically relevant serine- and metallo-carbapenemases.
Sci Rep, 10, 2020
|
|
3W77
 
 | | Crystal Structure of azoreductase AzrA | | 分子名称: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase | | 著者 | Ogata, D, Yu, J, Ooi, T, Yao, M. | | 登録日 | 2013-02-27 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3F5F
 
 | | Crystal structure of heparan sulfate 2-O-sulfotransferase from gallus gallus as a maltose binding protein fusion. | | 分子名称: | ADENOSINE-3'-5'-DIPHOSPHATE, Maltose-binding periplasmic protein, Heparan sulfate 2-O-sulfotransferase 1, ... | | 著者 | Bethea, H.N, Xu, D, Liu, J, Pedersen, L.C. | | 登録日 | 2008-11-03 | | 公開日 | 2008-12-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Redirecting the substrate specificity of heparan sulfate 2-O-sulfotransferase by structurally guided mutagenesis.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6TKI
 
 | | Tsetse thrombin inhibitor in complex with human alpha-thrombin - tetragonal form at 12.7keV | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, Thrombin heavy chain, ... | | 著者 | Calisto, B.M, Ripoll-Rozada, J, de Sanctis, D, Pereira, P.J.B. | | 登録日 | 2019-11-28 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Sulfotyrosine-Mediated Recognition of Human Thrombin by a Tsetse Fly Anticoagulant Mimics Physiological Substrates.
Cell Chem Biol, 28, 2021
|
|
8A9B
 
 | |
1F7Z
 
 | | RAT TRYPSINOGEN K15A COMPLEXED WITH BOVINE PANCREATIC TRYPSIN INHIBITOR | | 分子名称: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | 著者 | Pasternak, A, White, A, Jeffery, C.J, Medina, N, Cahoon, M, Ringe, D, Hedstrom, L. | | 登録日 | 2000-06-28 | | 公開日 | 2001-07-04 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The energetic cost of induced fit catalysis: Crystal structures of trypsinogen mutants with enhanced activity and inhibitor affinity.
Protein Sci., 10, 2001
|
|
3F85
 
 | |
3JD0
 
 | | Glutamate dehydrogenase in complex with GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, mitochondrial | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3JD4
 
 | | Glutamate dehydrogenase in complex with NADH and GTP, closed conformation | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GUANOSINE-5'-TRIPHOSPHATE, Glutamate dehydrogenase 1, ... | | 著者 | Borgnia, M.J, Banerjee, S, Merk, A, Matthies, D, Bartesaghi, A, Rao, P, Pierson, J, Earl, L.A, Falconieri, V, Subramaniam, S, Milne, J.L.S. | | 登録日 | 2016-03-28 | | 公開日 | 2016-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Using Cryo-EM to Map Small Ligands on Dynamic Metabolic Enzymes: Studies with Glutamate Dehydrogenase.
Mol.Pharmacol., 89, 2016
|
|
3WKU
 
 | | Crystal structure of the anaerobic DesB-gallate complex | | 分子名称: | 3,4,5-trihydroxybenzoic acid, FE (III) ION, Gallate dioxygenase | | 著者 | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | 登録日 | 2013-10-31 | | 公開日 | 2014-04-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
6TNH
 
 | | Deoxyguanylosuccinate synthase (DgsS) quaternary structure with AMPPcP, dGMP, Asp, Magnesium at 2.21 Angstrom resolution | | 分子名称: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ASPARTIC ACID, Adenylosuccinate synthetase, ... | | 著者 | Sleiman, D, Loc'h, J, Haouz, A, Kaminski, P.A. | | 登録日 | 2019-12-08 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | A third purine biosynthetic pathway encoded by aminoadenine-based viral DNA genomes.
Science, 372, 2021
|
|
3WVM
 
 | | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with stearic acid | | 分子名称: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | 著者 | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Matsumura, H, Murakami, S, Inoue, T, Murata, M. | | 登録日 | 2014-05-25 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (0.88 Å) | | 主引用文献 | Water-mediated recognition of simple alkyl chains by heart-type fatty-acid-binding protein.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8A9A
 
 | |
6TNT
 
 | | SUMOylated apoAPC/C with repositioned APC2 WHB domain | | 分子名称: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | 著者 | Barford, D, Yatskevich, S. | | 登録日 | 2019-12-10 | | 公開日 | 2021-01-13 | | 最終更新日 | 2021-07-28 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Molecular mechanisms of APC/C release from spindle assembly checkpoint inhibition by APC/C SUMOylation.
Cell Rep, 34, 2021
|
|
1F89
 
 | | Crystal structure of Saccharomyces cerevisiae Nit3, a member of branch 10 of the nitrilase superfamily | | 分子名称: | 32.5 KDA PROTEIN YLR351C | | 著者 | Kumaran, D, Eswaramoorthy, S, Studier, F.W, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2000-06-29 | | 公開日 | 2001-10-04 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of a putative CN hydrolase from yeast
Proteins, 52, 2003
|
|
6TKL
 
 | | Non-cleavable tsetse thrombin inhibitor in complex with human alpha-thrombin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Prothrombin, ... | | 著者 | Calisto, B.M, Ripoll-Rozada, J, de Sanctis, D, Pereira, P.J.B. | | 登録日 | 2019-11-28 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Sulfotyrosine-Mediated Recognition of Human Thrombin by a Tsetse Fly Anticoagulant Mimics Physiological Substrates.
Cell Chem Biol, 28, 2021
|
|
3K7A
 
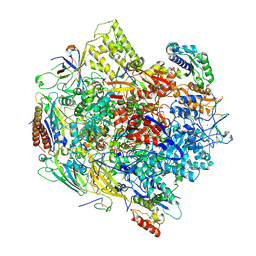 | | Crystal Structure of an RNA polymerase II-TFIIB complex | | 分子名称: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | 著者 | Liu, X, Bushnell, D.A, Wang, D, Calero, G, Kornberg, R.D. | | 登録日 | 2009-10-12 | | 公開日 | 2009-11-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism.
Science, 327, 2010
|
|
3WOD
 
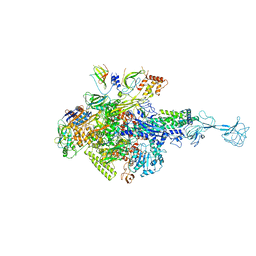 | | RNA polymerase-gp39 complex | | 分子名称: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | 著者 | Tagami, S, Sekine, S, Minakhin, L, Esyunina, D, Akasaka, R, Shirouzu, M, Kulbachinskiy, A, Severinov, K, Yokoyama, S. | | 登録日 | 2013-12-26 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Structural basis for promoter specificity switching of RNA polymerase by a phage factor.
Genes Dev., 28, 2014
|
|
3JRD
 
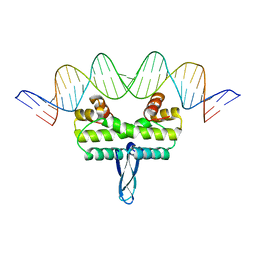 | |
8AYO
 
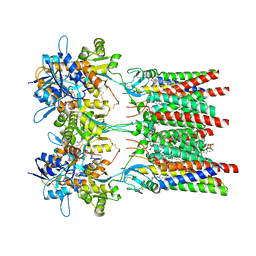 | | Open state GluA1/A2 AMPA receptor in complex with TARP gamma 8 and ligand JNJ-61432059 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 5-[2-(4-fluorophenyl)-7-(4-oxidanylpiperidin-1-yl)pyrazolo[1,5-c]pyrimidin-3-yl]-1,3-dihydroindol-2-one, ... | | 著者 | Zhang, D, Lape, R, Shaikh, S, Kohegyi, B, Watson, J.F, Cais, O, Nakagawa, T, Greger, I.H. | | 登録日 | 2022-09-02 | | 公開日 | 2023-04-19 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Modulatory mechanisms of TARP gamma 8-selective AMPA receptor therapeutics.
Nat Commun, 14, 2023
|
|
1F8F
 
 | | CRYSTAL STRUCTURE OF BENZYL ALCOHOL DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS | | 分子名称: | BENZYL ALCOHOL DEHYDROGENASE, ETHANOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Beauchamp, J.C, Gillooly, D, Warwicker, J, Fewson, C.A, Lapthorn, A.J. | | 登録日 | 2000-06-30 | | 公開日 | 2003-07-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of Benzyl Alcohol Dehydrogenase from Acinetobacter calcoaceticus
To be Published
|
|
8AYM
 
 | | Resting state GluA1/A2 AMPA receptor in complex with TARP gamma 8 and ligand JNJ-55511118 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 6-[2-chloro-6-(trifluoromethoxy)phenyl]-1H-benzimidazol-2-ol, ... | | 著者 | Zhang, D, Lape, R, Shaikh, S, Kohegyi, B, Watson, J.F, Cais, O, Nakagawa, T, Greger, I.H. | | 登録日 | 2022-09-02 | | 公開日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Modulatory mechanisms of TARP gamma 8-selective AMPA receptor therapeutics.
Nat Commun, 14, 2023
|
|
3CS3
 
 | | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis | | 分子名称: | GLYCEROL, SULFATE ION, Sugar-binding transcriptional regulator, ... | | 著者 | Patskovsky, Y, Romero, R, Freeman, J, Iizuka, M, Groshong, C, Smith, D, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-04-08 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structure of sugar-binding transcriptional regulator (LacI family) from Enterococcus faecalis.
To be Published
|
|
