8I21
 
 | | Cryo-EM structure of 6-subunit Smc5/6 arm region | | 分子名称: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | 著者 | Jun, Z, Qian, L, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | 登録日 | 2023-01-13 | | 公開日 | 2024-06-26 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (6.02 Å) | | 主引用文献 | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7F32
 
 | |
8UAB
 
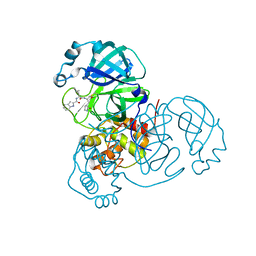 | | SARS-CoV-2 main protease (Mpro) complex with AC1115 | | 分子名称: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | 著者 | DuPrez, K.T, Chao, A, Han, F.Q. | | 登録日 | 2023-09-20 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.781 Å) | | 主引用文献 | Olgotrelvir, a dual inhibitor of SARS-CoV-2 M pro and cathepsin L, as a standalone antiviral oral intervention candidate for COVID-19
Med, 5, 2024
|
|
8UAC
 
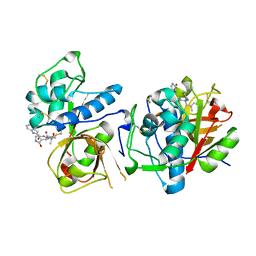 | | CATHEPSIN L IN COMPLEX WITH AC1115 | | 分子名称: | Cathepsin L, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | 著者 | Chao, A, DuPrez, K.T, Han, F.Q. | | 登録日 | 2023-09-20 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Olgotrelvir, a dual inhibitor of SARS-CoV-2 M pro and cathepsin L, as a standalone antiviral oral intervention candidate for COVID-19.
Med, 5, 2024
|
|
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | 分子名称: | CidA_I gamma/2 protein, CidB_I b/2 protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | 分子名称: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIU
 
 | | Crystal structure of the DUB domain of Wolbachia cytoplasmic incompatibility factor CidB from wMel | | 分子名称: | ULP_PROTEASE domain-containing protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIT
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wMel | | 分子名称: | bacteria factor 1 | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
6WHC
 
 | | CryoEM Structure of the glucagon receptor with a dual-agonist peptide | | 分子名称: | Dual-agonist peptide, Glucagon receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Belousoff, M.J, Sexton, P, Danev, R. | | 登録日 | 2020-04-07 | | 公開日 | 2020-05-27 | | 最終更新日 | 2025-06-04 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-electron microscopy structure of the glucagon receptor with a dual-agonist peptide.
J.Biol.Chem., 295, 2020
|
|
7RN0
 
 | |
7RN1
 
 | |
8HJU
 
 | | Cryo-EM structure of native RC-LH complex from Roseiflexus castenholzii at 10,000 lux | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-methyl-3-[(2E,6E,10E,14E,18E,22E,26E,30E,34E,38E)-3,7,11,15,19,23,27,31,35,39,43-undecamethyltetratetraconta-2,6,10,1 4,18,22,26,30,34,38,42-undecaen-1-yl]naphthalene-1,4-dione, Alpha subunit of light-harvesting 1, ... | | 著者 | Xu, X, Xin, J. | | 登録日 | 2022-11-23 | | 公開日 | 2023-09-20 | | 最終更新日 | 2024-10-30 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Carotenoid assembly regulates quinone diffusion and the Roseiflexus castenholzii reaction center-light harvesting complex architecture.
Elife, 12, 2023
|
|
5D6P
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-[4-(hydroxymethyl)-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl]urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-12 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
3CP9
 
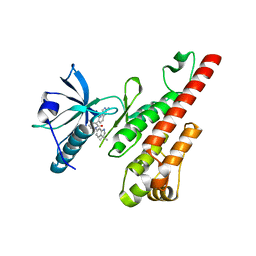 | | Crystal structure of the VEGFR2 kinase domain in complex with a pyridone inhibitor | | 分子名称: | 3-(2-aminoquinazolin-6-yl)-1-(3,3-dimethylindolin-6-yl)-4-methylpyridin-2(1H)-one, Vascular endothelial growth factor receptor 2 | | 著者 | Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | 登録日 | 2008-03-31 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Discovery of Aryl Aminoquinazoline Pyridones as Potent, Selective, and Orally Efficacious Inhibitors of Receptor Tyrosine Kinase c-Kit.
J.Med.Chem., 51, 2008
|
|
3J2Y
 
 | |
3CPB
 
 | | Crystal structure of the VEGFR2 kinase domain in complex with a bisamide inhibitor | | 分子名称: | N'-(6-aminopyridin-3-yl)-N-(2-cyclopentylethyl)-4-methyl-benzene-1,3-dicarboxamide, Vascular endothelial growth factor receptor 2 | | 著者 | Whittington, D.A, Long, A.M, Rose, P, Gu, Y, Zhao, H. | | 登録日 | 2008-03-31 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of Aryl Aminoquinazoline Pyridones as Potent, Selective, and Orally Efficacious Inhibitors of Receptor Tyrosine Kinase c-Kit.
J.Med.Chem., 51, 2008
|
|
3J2W
 
 | |
8HZ5
 
 | |
8HZ4
 
 | |
5D6Q
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a ligand | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-ethyl-3-{4-[(E)-2-(pyridin-3-yl)ethenyl]-5-(1H-pyrrol-2-yl)-1,3-thiazol-2-yl}urea, DNA gyrase subunit B, ... | | 著者 | Zhang, J, Yang, Q, Cross, J.B, Romero, J.A.C, Ryan, M.D, Lippa, B, Dolle, R.E, Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C. | | 登録日 | 2015-08-12 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery of Azaindole Ureas as a Novel Class of Bacterial Gyrase B Inhibitors.
J.Med.Chem., 58, 2015
|
|
6MDU
 
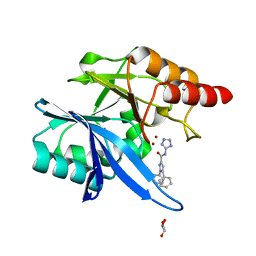 | | Crystal structure of NDM-1 with compound 7 | | 分子名称: | 1,5-diphenyl-N-(1H-tetrazol-5-yl)-1H-pyrazole-3-carboxamide, GLYCEROL, Metallo-beta-lactamase type 2, ... | | 著者 | Akhtar, A, Chen, Y. | | 登録日 | 2018-09-05 | | 公開日 | 2019-04-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
8HVS
 
 | |
5X6O
 
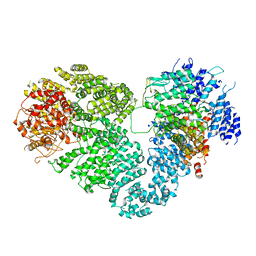 | | Intact ATR/Mec1-ATRIP/Ddc2 complex | | 分子名称: | DNA damage checkpoint protein LCD1, Serine/threonine-protein kinase MEC1 | | 著者 | Wang, X, Ran, T, Cai, G. | | 登録日 | 2017-02-22 | | 公開日 | 2017-12-20 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | 3.9 angstrom structure of the yeast Mec1-Ddc2 complex, a homolog of human ATR-ATRIP.
Science, 358, 2017
|
|
7FAT
 
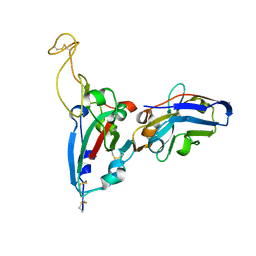 | | Structure Determination of the RBD-NB1A7 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, nb_1A7 | | 著者 | Geng, Y, Shi, Z.Z, Li, X.Y, Wang, L, Sun, Z.C, Zhang, H.W, Chen, X.C. | | 登録日 | 2021-07-07 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural basis of nanobodies neutralizing SARS-CoV-2 variants
Structure, 30, 2022
|
|
4LN5
 
 | | Crystal structure of a trap periplasmic solute binding protein from burkholderia ambifaria (Bamb_6123), TARGET EFI-510059, with bound glycerol and chloride ion | | 分子名称: | CHLORIDE ION, GLYCEROL, TRAP dicarboxylate transporter, ... | | 著者 | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2013-07-11 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
