3WUE
 
 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 | | 分子名称: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | 著者 | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y, Guo, R.T. | | 登録日 | 2014-04-23 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WQN
 
 | | Crystal structure of Rv3378c_Y51F with TPP | | 分子名称: | (2E)-3-methyl-5-[(1R,2S,8aS)-1,2,5,5-tetramethyl-1,2,3,5,6,7,8,8a-octahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Diterpene synthase, PHOSPHATE ION | | 著者 | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | 登録日 | 2014-01-28 | | 公開日 | 2014-02-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
5JBY
 
 | |
5JBV
 
 | |
5J8P
 
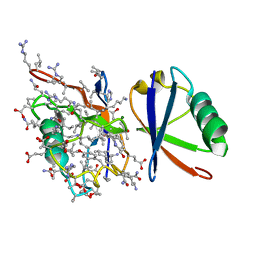 | |
3WDJ
 
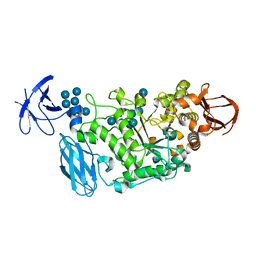 | | Crystal structure of Pullulanase complexed with maltotetraose from Anoxybacillus sp. LM18-11 | | 分子名称: | CALCIUM ION, Type I pullulanase, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Xu, J, Ren, F, Huang, C.H, Zheng, Y, Zhen, J, Ko, T.P, Chen, C.C, Chan, H.C, Guo, R.T, Ma, Y, Song, H. | | 登録日 | 2013-06-18 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Cloning, Expression, Functional and Structural Studies of Pullulanase from Anoxybacillus sp. LM18-11
To be Published
|
|
3WQL
 
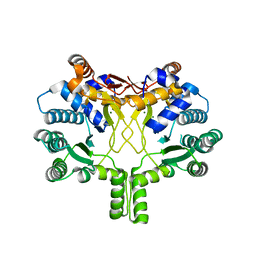 | | Crystal structure of Rv3378c with Mg2+ and PPi | | 分子名称: | Diterpene synthase, MAGNESIUM ION | | 著者 | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | 登録日 | 2014-01-28 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
5DXK
 
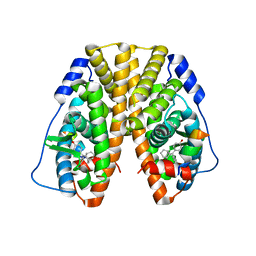 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-[(9s)-bicyclo[3.3.1]non-9-ylmethanediyl]diphenol | | 分子名称: | 4,4'-[(9s)-bicyclo[3.3.1]non-9-ylmethanediyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-23 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.226 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DY8
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[(3S)-3-ethylcyclohexylidene]methanediyl}diphenol | | 分子名称: | 4,4'-{[(3S)-3-ethylcyclohexylidene]methanediyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-24 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.031 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DZH
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[4-(2-hydroxyethyl)cyclohexylidene]methanediyl}diphenol | | 分子名称: | 4,4'-{[4-(2-hydroxyethyl)cyclohexylidene]methanediyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-25 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DWG
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Triaryl-substituted Imine Analog, 4-{(E)-(4-hydroxyphenyl)[(2-methylphenyl)imino]methyl}benzene-1,3-diol | | 分子名称: | 4-{(E)-(4-hydroxyphenyl)[(2-methylphenyl)imino]methyl}benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-22 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5E15
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[4-(2-hydroxyethyl)cyclohexylidene]methanediyl}diphenol | | 分子名称: | 4,4'-({4-[2-(4-fluorobutoxy)ethyl]cyclohexylidene}methanediyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-29 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DUE
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with a para-Hydroxyl-substituted, Sulfoxide-bridged Oxabicyclic Heptene Sulfonate (SOBHS)-2 Analog 4-hydroxyphenyl (1S,2S,4S,5S,6R,7S)-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]heptane-2-sulfonate 7-oxide | | 分子名称: | 4-hydroxyphenyl (1S,2S,4S,7S)-5,6-bis(4-hydroxy-2-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate 7-oxide, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-18 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DUO
 
 | | Crystal structure of native translocator protein 18kDa (TSPO) from Rhodobacter sphaeroides (A139T Mutant) in C2 space group | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FORMIC ACID, PROTOPORPHYRIN IX, ... | | 著者 | Li, F, Liu, J, Zheng, Y, Garavito, R.M, Ferguson-Miller, S. | | 登録日 | 2015-09-20 | | 公開日 | 2015-11-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Response to Comment on "Crystal structures of translocator protein (TSPO) and mutant mimic of a human polymorphism".
Science, 350, 2015
|
|
5DKG
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a t-butyl-substituted, methyl, triaryl-ethylene derivative 4,4'-[2-(4-tert-butylphenyl)prop-1-ene-1,1-diyl]diphenol | | 分子名称: | 4,4'-[2-(4-tert-butylphenyl)prop-1-ene-1,1-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-03 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DMC
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a nitrile-substituted triaryl-ethylene derivative 3,3-bis(4-hydroxyphenyl)-2-phenylprop-2-enenitrile | | 分子名称: | 3,3-bis(4-hydroxyphenyl)-2-phenylprop-2-enenitrile, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-08 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.403 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DVV
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with a Triaryl-imine analog 4,4'-(phenylcarbonimidoyl)diphenol | | 分子名称: | 4,4'-(phenylcarbonimidoyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-21 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.505 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DXQ
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-[(1s,5s)-bicyclo[3.3.1]non-9-ylidenemethanediyl]diphenol | | 分子名称: | 4,4'-[(1s,5s)-bicyclo[3.3.1]non-9-ylidenemethanediyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-23 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5E19
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative methyl {4-[bis(4-hydroxyphenyl)methylidene]cyclohexyl}acetate | | 分子名称: | Estrogen receptor, Nuclear receptor coactivator 2, methyl {4-[bis(4-hydroxyphenyl)methylidene]cyclohexyl}acetate | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-29 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DKS
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 2-naphthylamino-substituted, ethyl, triaryl-ethylene derivative 4,4'-{2-[3-(naphthalen-1-ylamino)phenyl]but-1-ene-1,1-diyl}diphenol | | 分子名称: | 4,4'-{2-[3-(naphthalen-1-ylamino)phenyl]but-1-ene-1,1-diyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-03 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DLR
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a triaryl-ethylene compound 4,4'-(2-phenylethene-1,1-diyl)diphenol | | 分子名称: | 4,4'-(2-phenylethene-1,1-diyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-07 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DK9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a phenylamino-substituted ethyl triaryl-ethylene derivative 4,4'-{2-[3-(phenylamino)phenyl]but-1-ene-1,1-diyl}diphenol | | 分子名称: | 4,4'-{2-[3-(phenylamino)phenyl]but-1-ene-1,1-diyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-03 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DXR
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-{[(3R)-3-methylcyclohexylidene]methanediyl}diphenol | | 分子名称: | 4,4'-{[(3R)-3-methylcyclohexylidene]methanediyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-23 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DYB
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-(3,4-dihydronaphthalen-2(1H)-ylidenemethanediyl)diphenol | | 分子名称: | 4,4'-(3,4-dihydronaphthalen-2(1H)-ylidenemethanediyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-24 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DP0
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 4-fluorophenylamino-substituted triaryl-ethylene derivative 4,4'-(2-{3-[(4-fluorophenyl)amino]phenyl}ethene-1,1-diyl)diphenol | | 分子名称: | 4,4'-(2-{3-[(4-fluorophenyl)amino]phenyl}ethene-1,1-diyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | 登録日 | 2015-09-11 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.382 Å) | | 主引用文献 | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
