5YKZ
 
 | | The crystal structure of Penaeus vannamei nodavirus P-domain (P21) | | 分子名称: | Capsid protein | | 著者 | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | 登録日 | 2017-10-16 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKV
 
 | | The crystal structure of Macrobrachium rosenbergii nodavirus P-domain | | 分子名称: | Capsid protein | | 著者 | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | 登録日 | 2017-10-16 | | 公開日 | 2018-10-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YKU
 
 | | The crystal structure of Macrobrachium rosenbergii nodavirus P-domain with Zn ions | | 分子名称: | Capsid protein, ZINC ION | | 著者 | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | 登録日 | 2017-10-16 | | 公開日 | 2018-10-24 | | 最終更新日 | 2019-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
5YL1
 
 | | T=1 subviral particle of Penaeus vannamei nodavirus capsid protein deletion mutant (delta 1-37 & 251-368) | | 分子名称: | CALCIUM ION, Capsid protein | | 著者 | Chen, N.C, Yoshimura, M, Lin, C.C, Guan, H.H, Chuankhayan, P, Chen, C.J. | | 登録日 | 2017-10-16 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.12 Å) | | 主引用文献 | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
3UZB
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | 分子名称: | 2-OXO-4-METHYLPENTANOIC ACID, Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | 登録日 | 2011-12-07 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
7EJV
 
 | | The co-crystal structure of DYRK2 with YK-2-69 | | 分子名称: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [6-[[4-[2-(dimethylamino)-1,3-benzothiazol-6-yl]-5-fluoranyl-pyrimidin-2-yl]amino]pyridin-3-yl]-(4-ethylpiperazin-1-yl)methanone | | 著者 | Li, Z, Xiao, Y, Yuan, K, Kuang, W, Xiuquan, Y, Yang, P. | | 登録日 | 2021-04-02 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Targeting dual-specificity tyrosine phosphorylation-regulated kinase 2 with a highly selective inhibitor for the treatment of prostate cancer.
Nat Commun, 13, 2022
|
|
3V1Y
 
 | | Crystal structures of glyceraldehyde-3-phosphate dehydrogenase complexes with NAD | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Tien, Y.C, Chuankhayan, P, Lin, Y.H, Chang, S.L, Chen, C.J. | | 登録日 | 2011-12-10 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis
Plant Mol.Biol., 80, 2012
|
|
3UYY
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | 分子名称: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | 登録日 | 2011-12-07 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
3UZO
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | 分子名称: | Branched-chain-amino-acid aminotransferase, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | 登録日 | 2011-12-07 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
2KJU
 
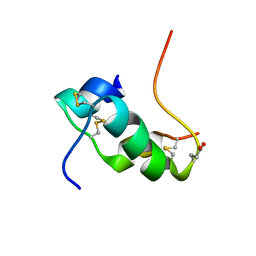 | | NMR structure of human insulin mutant glu-b21-d-glu, his-b10 asp pro-b28-lys, lys-b29-pro, 20 structures | | 分子名称: | Insulin | | 著者 | Hua, Q.X, Huang, K, Hu, S.Q, Katsoyanni, P, Weiss, M.A. | | 登録日 | 2009-06-10 | | 公開日 | 2010-06-16 | | 最終更新日 | 2021-10-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Acceleration of Protein Fibrillation by Chiral Destabilization of Beta-Turn
To be Published
|
|
7XPE
 
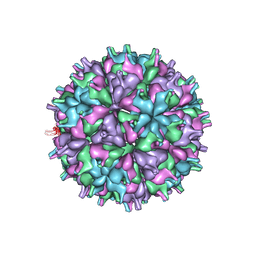 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 8.5 | | 分子名称: | Capsid protein alpha | | 著者 | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | 登録日 | 2022-05-04 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.32 Å) | | 主引用文献 | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7XGZ
 
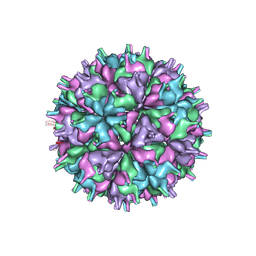 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 7.5 | | 分子名称: | Capsid protein alpha | | 著者 | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | 登録日 | 2022-04-07 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.24 Å) | | 主引用文献 | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions
Nat Commun, 14, 2023
|
|
7XPB
 
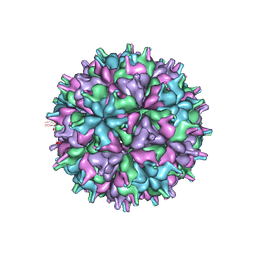 | | Cryo-EM structure of the T=4 lake sinai virus 2 virus-like capsid at pH 6.5 | | 分子名称: | Capsid protein alpha | | 著者 | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | 登録日 | 2022-05-04 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.91 Å) | | 主引用文献 | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7XPA
 
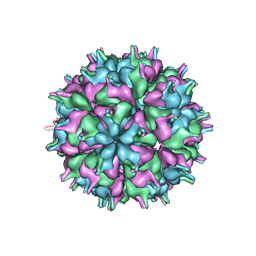 | | Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 7.5 | | 分子名称: | Capsid protein alpha | | 著者 | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | 登録日 | 2022-05-04 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7XPG
 
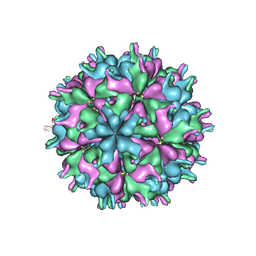 | | Cryo-EM structure of the T=3 lake sinai virus 1 (delta-N48) virus-like capsid at pH 6.5 | | 分子名称: | Capsid protein alpha, RNA (5'-R(P*UP*G)-3') | | 著者 | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | 登録日 | 2022-05-04 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.51 Å) | | 主引用文献 | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7XPD
 
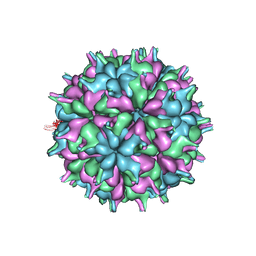 | | Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 6.5 | | 分子名称: | Capsid protein alpha | | 著者 | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | 登録日 | 2022-05-04 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.74 Å) | | 主引用文献 | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7XPF
 
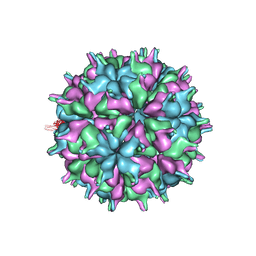 | | Cryo-EM structure of the T=3 lake sinai virus 2 virus-like capsid at pH 8.5 | | 分子名称: | Capsid protein alpha | | 著者 | Chen, N.C, Wang, C.H, Chen, C.J, Yoshimura, M, Guan, H.H, Chuankhayan, P, Lin, C.C. | | 登録日 | 2022-05-04 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Structures of honeybee-infecting Lake Sinai virus reveal domain functions and capsid assembly with dynamic motions.
Nat Commun, 14, 2023
|
|
7VUN
 
 | | Design, modification, evaluation and cocrystal studies of novel phthalimides regulating PD-1/PD-L1 interaction | | 分子名称: | (2~{S},3~{S})-2-[[6-[(3-cyanophenyl)methoxy]-2-(2-methyl-3-phenyl-phenyl)-1,3-bis(oxidanylidene)isoindol-5-yl]methylamino]-3-oxidanyl-butanoic acid, Programmed cell death 1 ligand 1 | | 著者 | Cheng, Y, Sun, C.L, Chen, M.R, Yang, P, Xiao, Y.B. | | 登録日 | 2021-11-03 | | 公開日 | 2022-09-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Novel phthalimides regulating PD-1/PD-L1 interaction as potential immunotherapy agents.
Acta Pharm Sin B, 12, 2022
|
|
7XE4
 
 | | structure of a membrane-bound glycosyltransferase | | 分子名称: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | 登録日 | 2022-03-29 | | 公開日 | 2023-03-29 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
4DNW
 
 | | Crystal structure of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.773 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNV
 
 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
4DNU
 
 | | Crystal structure of the W285A mutant of UVB-resistance protein UVR8 | | 分子名称: | AT5g63860/MGI19_6 | | 著者 | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | 登録日 | 2012-02-09 | | 公開日 | 2012-03-07 | | 最終更新日 | 2013-07-17 | | 実験手法 | X-RAY DIFFRACTION (1.764 Å) | | 主引用文献 | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
3O2F
 
 | |
3O0I
 
 | |
7XZO
 
 | | Formate-tetrahydrofolate ligase in complex with ATP | | 分子名称: | (R,R)-2,3-BUTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Formate--tetrahydrofolate ligase, ... | | 著者 | Fang, C.L, Zhang, Y. | | 登録日 | 2022-06-03 | | 公開日 | 2023-06-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.31 Å) | | 主引用文献 | Identification of FtfL as a novel target of berberine in intestinal bacteria.
Bmc Biol., 21, 2023
|
|
