1HF0
 
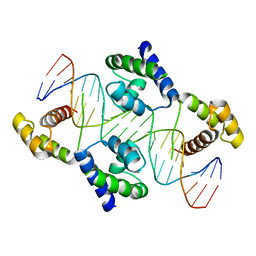 | | Crystal structure of the DNA-binding domain of Oct-1 bound to DNA as a dimer | | 分子名称: | DNA 5'-D(*CP*AP*CP*AP*TP*TP*TP*GP*AP*AP*AP*GP*GP* CP*AP*AP*AP*TP*GP*GP*AP*G)-3', DNA 5'-D(*CP*TP*CP*CP*AP*TP*TP*TP*GP*CP*CP*TP*TP* TP*CP*AP*AP*AP*TP*GP*TP*G)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1 | | 著者 | Remenyi, A, Tomilin, A, Pohl, E, Scholer, H.R, Wilmanns, M. | | 登録日 | 2000-11-27 | | 公開日 | 2001-11-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping
Mol.Cell, 8, 2001
|
|
1GT0
 
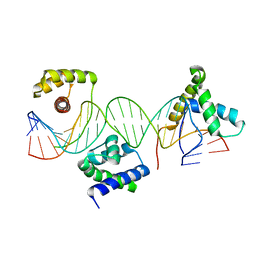 | | Crystal structure of a POU/HMG/DNA ternary complex | | 分子名称: | 5'-D(*AP*TP*CP*CP*CP*AP*TP*TP*AP*GP* CP*AP*TP*CP*CP*AP*AP*AP*CP*AP*AP*AP*GP*A)-3', 5'-D(*TP*TP*CP*TP*TP*TP*GP*TP*TP*TP* GP*GP*AP* TP*GP*CP*TP*AP*AP*TP*GP*GP*GP*A)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1, ... | | 著者 | Remenyi, A, Wilmanns, M. | | 登録日 | 2002-01-09 | | 公開日 | 2003-01-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal Structure of a POU/Hmg/DNA Ternary Complex Suggests Differential Assembly of Oct4 and Sox2 on Two Enhancers
Genes Dev., 17, 2003
|
|
1TKI
 
 | |
6FHB
 
 | |
6FHA
 
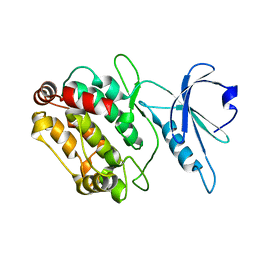 | |
6FWN
 
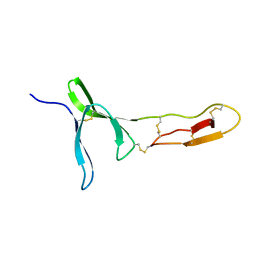 | | Structure and dynamics of the platelet integrin-binding C4 domain of von Willebrand factor | | 分子名称: | von Willebrand factor | | 著者 | Xu, E.-R, von Buelow, S, Chen, P.-C, Lenting, P.J, Kolsek, K, Aponte-Santamaria, C, Simon, B, Foot, J, Obser, T, Graeter, F, Schneppenheim, R, Denis, C.V, Wilmanns, M, Hennig, J. | | 登録日 | 2018-03-06 | | 公開日 | 2018-10-24 | | 最終更新日 | 2019-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and dynamics of the platelet integrin-binding C4 domain of von Willebrand factor.
Blood, 133, 2019
|
|
1KY8
 
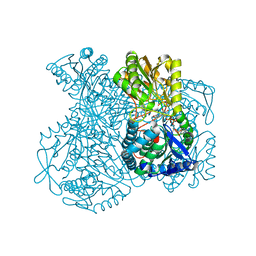 | | Crystal Structure of the Non-phosphorylating glyceraldehyde-3-phosphate Dehydrogenase | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SODIUM ION, glyceraldehyde-3-phosphate dehydrogenase | | 著者 | Pohl, E, Brunner, N, Wilmanns, M, Hensel, R. | | 登録日 | 2002-02-04 | | 公開日 | 2003-02-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The Crystal Structure of the Allosteric Non-phosphorylating
glyceraldehyde-3-phosphate Dehydrogenase from the Hyperthermophilic
Archaeum Thermoproteus tenax
J.Biol.Chem., 277, 2002
|
|
6H4L
 
 | | Structure of Titin M4 trigonal form | | 分子名称: | CHLORIDE ION, Titin, ZINC ION | | 著者 | Sauer, F, Wilmanns, M. | | 登録日 | 2018-07-21 | | 公開日 | 2019-08-07 | | 最終更新日 | 2020-02-19 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural diversity in the atomic resolution 3D fingerprint of the titin M-band segment.
Plos One, 14, 2019
|
|
1JO8
 
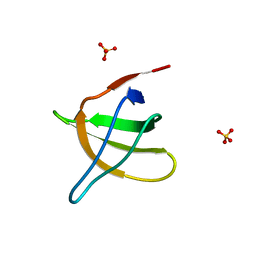 | | Structural analysis of the yeast actin binding protein Abp1 SH3 domain | | 分子名称: | ACTIN BINDING PROTEIN, SULFATE ION | | 著者 | Fazi, B, Cope, M.J, Douangamath, A, Ferracuti, S, Schirwitz, K, Zucconi, A, Drubin, D.G, Wilmanns, M, Cesareni, G, Castagnoli, L. | | 登録日 | 2001-07-27 | | 公開日 | 2002-03-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Unusual binding properties of the SH3 domain of the yeast actin-binding protein Abp1: structural and functional analysis.
J.Biol.Chem., 277, 2002
|
|
4I8A
 
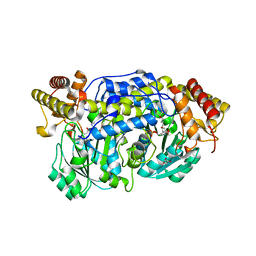 | | Alanine-glyoxylate aminotransferase variant S187F | | 分子名称: | GLYCEROL, Serine-pyruvate aminotransferase | | 著者 | Fodor, K, Oppici, E, Williams, C, Cellini, B, Wilmanns, M. | | 登録日 | 2012-12-03 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of the S187F variant of human liver alanine: Aminotransferase associated with primary hyperoxaluria type I and its functional implications.
Proteins, 81, 2013
|
|
2O31
 
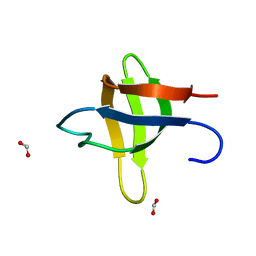 | |
2O9S
 
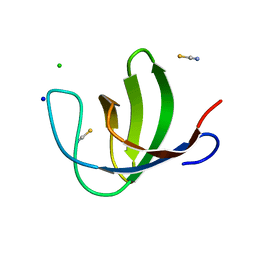 | |
2WL8
 
 | | X-ray crystal structure of Pex19p | | 分子名称: | PEROXISOMAL BIOGENESIS FACTOR 19 | | 著者 | Schueller, N, Holton, S.J, Stanley, W.A, Song, Y.H, Konarev, P, Roessle, M, Erdmann, R, Schliebs, W, Wilmanns, M. | | 登録日 | 2009-06-22 | | 公開日 | 2010-06-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | The Peroxisomal Receptor Pex19P Forms a Helical Mpts Recognition Domain.
Embo J., 29, 2010
|
|
2O9V
 
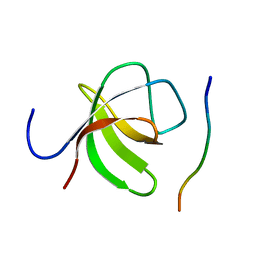 | |
1OOT
 
 | |
1QDL
 
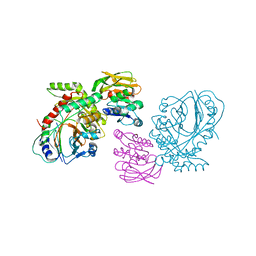 | | THE CRYSTAL STRUCTURE OF ANTHRANILATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS | | 分子名称: | PROTEIN (ANTHRANILATE SYNTHASE (TRPE-SUBUNIT)), PROTEIN (ANTHRANILATE SYNTHASE (TRPG-SUBUNIT)) | | 著者 | Knoechel, T, Ivens, A, Hester, G, Gonzalez, A, Bauerle, R, Wilmanns, M, Kirschner, K, Jansonius, J.N. | | 登録日 | 1999-05-20 | | 公開日 | 1999-08-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of anthranilate synthase from Sulfolobus solfataricus: functional implications.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2NNY
 
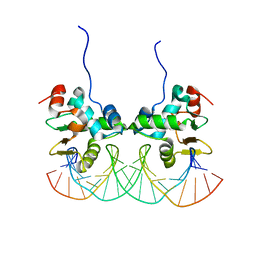 | | Crystal structure of the Ets1 dimer DNA complex. | | 分子名称: | 5'-D(*A*CP*TP*CP*CP*AP*GP*GP*AP*AP*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*TP*CP*T)-3', 5'-D(*T*AP*GP*AP*CP*AP*GP*GP*AP*AP*GP*CP*AP*CP*TP*TP*CP*CP*TP*GP*GP*AP*G)-3', C-ets-1 protein | | 著者 | Lamber, E.P, Kachalova, G.S, Wilmanns, M. | | 登録日 | 2006-10-24 | | 公開日 | 2008-03-04 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Regulation of the transcription factor Ets-1 by DNA-mediated homo-dimerization.
Embo J., 27, 2008
|
|
1PWT
 
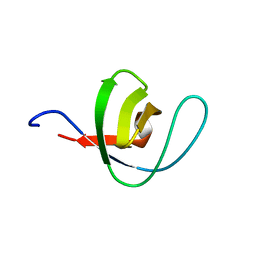 | | THERMODYNAMIC ANALYSIS OF ALPHA-SPECTRIN SH3 AND TWO OF ITS CIRCULAR PERMUTANTS WITH DIFFERENT LOOP LENGTHS: DISCERNING THE REASONS FOR RAPID FOLDING IN PROTEINS | | 分子名称: | ALPHA SPECTRIN | | 著者 | Martinez, J.C, Viguera, A.R, Berisio, R, Wilmanns, M, Mateo, P.L, Filmonov, V.V, Serrano, L. | | 登録日 | 1998-10-06 | | 公開日 | 1999-05-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Thermodynamic analysis of alpha-spectrin SH3 and two of its circular permutants with different loop lengths: discerning the reasons for rapid folding in proteins.
Biochemistry, 38, 1999
|
|
1RNJ
 
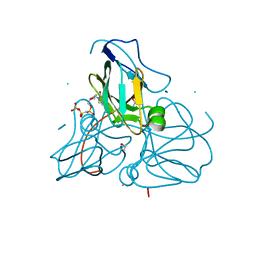 | | Crystal structure of inactive mutant dUTPase complexed with substrate analogue imido-dUTP | | 分子名称: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | 著者 | Barabas, O, Pongracz, V, Kovari, J, Wilmanns, M, Vertessy, B.G. | | 登録日 | 2003-12-01 | | 公開日 | 2004-09-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Insights into the Catalytic Mechanism of Phosphate Ester Hydrolysis by dUTPase.
J.Biol.Chem., 279, 2004
|
|
4KYO
 
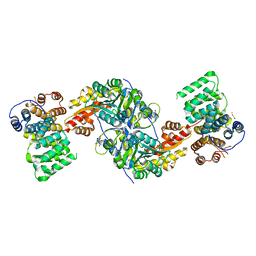 | | Alanine-glyoxylate aminotransferase variant K390A in complex with the TPR domain of human Pex5p | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, Peroxisomal targeting signal 1 receptor, ... | | 著者 | Fodor, K, Lou, Y, Wilmanns, M. | | 登録日 | 2013-05-29 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Ligand-Induced Compaction of the PEX5 Receptor-Binding Cavity Impacts Protein Import Efficiency into Peroxisomes.
Traffic, 16, 2015
|
|
4KXK
 
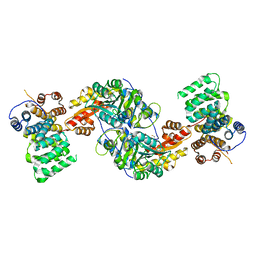 | | Alanine-glyoxylate aminotransferase variant K390A/K391A in complex with the TPR domain of human Pex5p | | 分子名称: | BETA-MERCAPTOETHANOL, Peroxisomal targeting signal 1 receptor, SULFATE ION, ... | | 著者 | Fodor, K, Lou, Y, Wilmanns, M. | | 登録日 | 2013-05-27 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Ligand-Induced Compaction of the PEX5 Receptor-Binding Cavity Impacts Protein Import Efficiency into Peroxisomes.
Traffic, 16, 2015
|
|
2W85
 
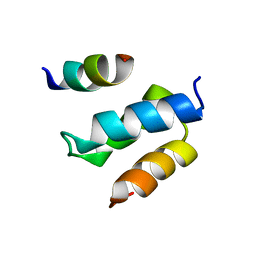 | | Structure of Pex14 in complex with Pex19 | | 分子名称: | PEROXIN-19, PEROXISOMAL MEMBRANE ANCHOR PROTEIN PEX14 | | 著者 | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | 登録日 | 2009-01-09 | | 公開日 | 2009-02-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for Competitive Interactions of Pex14 with the Import Receptors Pex5 and Pex19.
Embo J., 28, 2009
|
|
3ZXT
 
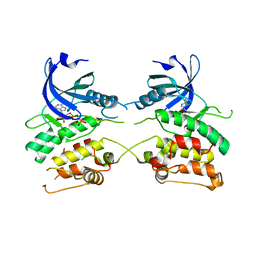 | |
3ZS4
 
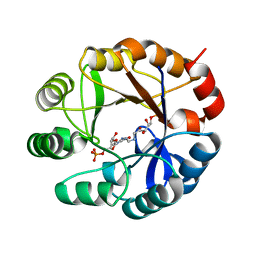 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS PHOSPHORIBOSYL ISOMERASE WITH BOUND PRFAR | | 分子名称: | PHOSPHORIBOSYL ISOMERASE A, PHOSPHORIC ACID MONO-[5-({[5-CARBAMOYL-3-(5-PHOSPHONOOXY-5-DEOXY-RIBOFURANOSYL)- 3H-IMIDAZOL-4-YLAMINO]-METHYL}-AMINO)-2,3,4-TRIHYDROXY-PENTYL] ESTER | | 著者 | Due, A.V, Kuper, J, Geerlof, A, Wilmanns, M. | | 登録日 | 2011-06-22 | | 公開日 | 2012-07-11 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of Mycobacterium Tuberculosis Phosphoribosyl Isomerase with Bound Prfar
To be Published
|
|
2W84
 
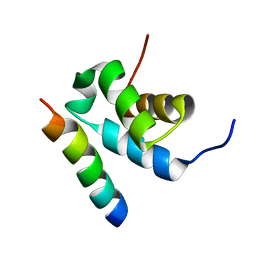 | | Structure of Pex14 in complex with Pex5 | | 分子名称: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | 著者 | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | 登録日 | 2009-01-09 | | 公開日 | 2009-02-17 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19.
EMBO J., 28, 2009
|
|
