6PW9
 
 | | Cryo-EM structure of human NatE/HYPK complex | | 分子名称: | ACETYL COENZYME *A, Huntingtin-interacting protein K, INOSITOL HEXAKISPHOSPHATE, ... | | 著者 | Deng, S, Marmorstein, R. | | 登録日 | 2019-07-22 | | 公開日 | 2020-02-19 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (4.03 Å) | | 主引用文献 | Molecular basis for N-terminal acetylation by human NatE and its modulation by HYPK.
Nat Commun, 11, 2020
|
|
6ILG
 
 | |
6ILF
 
 | |
6ILE
 
 | | CRYSTAL STRUCTURE OF A MUTANT PTAL-N*01:01 FOR 2.9 ANGSTROM, 52M 53D 54L DELETED | | 分子名称: | Beta-2-microglobulin, HEV-1, MHC class I antigen | | 著者 | Qu, Z.H, Zhang, N.Z, Xia, C. | | 登録日 | 2018-10-17 | | 公開日 | 2019-07-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure and Peptidome of the Bat MHC Class I Molecule Reveal a Novel Mechanism Leading to High-Affinity Peptide Binding.
J Immunol., 202, 2019
|
|
6ILC
 
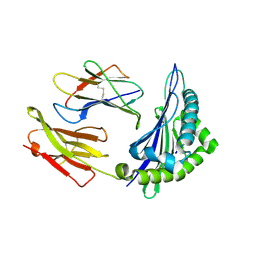 | |
7WKX
 
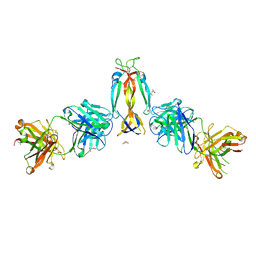 | | IL-17A in complex with the humanized antibody HB0017 | | 分子名称: | ACETIC ACID, Heavy chain of HB0017 Fab, Interleukin-17A, ... | | 著者 | Xu, J, Zhu, X, He, Y. | | 登録日 | 2022-01-12 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Structural and functional insights into a novel pre-clinical-stage antibody targeting IL-17A for treatment of autoimmune diseases.
Int.J.Biol.Macromol., 202, 2022
|
|
3TPX
 
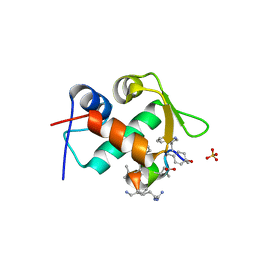 | |
5W0Q
 
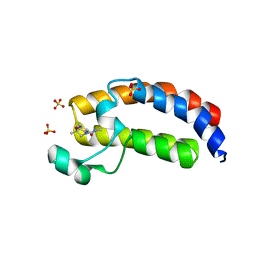 | | CREBBP Bromodomain in complex with Cpd17 (N,2,7-trimethyl-2,3-dihydro-4H-benzo[b][1,4]oxazine-4-carboxamide) | | 分子名称: | (2R)-N,2,7-trimethyl-2,3-dihydro-4H-1,4-benzoxazine-4-carboxamide, CREB-binding protein, SULFATE ION | | 著者 | Murray, J.M. | | 登録日 | 2017-05-31 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
5W0F
 
 | | CREBBP Bromodomain in complex with Cpd3 ((S)-1-(3-(6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | 分子名称: | 1-{3-[6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | 著者 | Murray, J.M. | | 登録日 | 2017-05-30 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
5W0L
 
 | | CREBBP Bromodomain in complex with Cpd10 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | 分子名称: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | 著者 | Murray, J.M. | | 登録日 | 2017-05-31 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.549 Å) | | 主引用文献 | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
5W0I
 
 | | CREBBP Bromodomain in complex with Cpd8 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | 分子名称: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | 著者 | Murray, J.M. | | 登録日 | 2017-05-30 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.43 Å) | | 主引用文献 | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
5YSX
 
 | |
6O07
 
 | |
3UIM
 
 | |
6A73
 
 | | Complex structure of CSN2 with IP6 | | 分子名称: | COP9 signalosome complex subunit 2,Endolysin, INOSITOL HEXAKISPHOSPHATE, SULFATE ION | | 著者 | Liu, L, Li, D, Rao, F, Wang, T. | | 登録日 | 2018-07-02 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.447 Å) | | 主引用文献 | Basis for metabolite-dependent Cullin-RING ligase deneddylation by the COP9 signalosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5KA8
 
 | | Protein Tyrosine Phosphatase 1B L192A mutant, open state | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Tyrosine-protein phosphatase non-receptor type 1 | | 著者 | Choy, M.S, Peti, W, Page, R. | | 登録日 | 2016-06-01 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.971 Å) | | 主引用文献 | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
5KA9
 
 | | Protein Tyrosine Phosphatase 1B L192A mutant in complex with TCS401, open state | | 分子名称: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Choy, M.S, Peti, W, Page, R. | | 登録日 | 2016-06-01 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
5KA0
 
 | | Protein Tyrosine Phosphatase 1B Delta helix 7, open state | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Choy, M.S, Peti, W, Page, R. | | 登録日 | 2016-06-01 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.991 Å) | | 主引用文献 | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
5KAD
 
 | | Protein Tyrosine Phosphatase 1B N193A mutant in complex with TCS401, closed state | | 分子名称: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Choy, M.S, Peti, W, Page, R. | | 登録日 | 2016-06-01 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
5K9W
 
 | | Protein Tyrosine Phosphatase 1B (1-301) in complex with TCS401, closed state | | 分子名称: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Choy, M.S, Peti, W, Page, R. | | 登録日 | 2016-06-01 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
5KA7
 
 | | Protein Tyrosine Phosphatase 1B T178A mutant in complex with TCS401, closed state | | 分子名称: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | 著者 | Choy, M.S, Peti, W, Page, R. | | 登録日 | 2016-06-01 | | 公開日 | 2017-03-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.061 Å) | | 主引用文献 | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
5K9V
 
 | | Protein Tyrosine Phosphatase 1B (1-301), open state | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Choy, M.S, Peti, W, Page, R. | | 登録日 | 2016-06-01 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
5KAA
 
 | | Protein Tyrosine Phosphatase 1B Delta helix 7, P185G mutant, open state | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Choy, M.S, Machado, L.E.S.F, Peti, W, Page, R. | | 登録日 | 2016-06-01 | | 公開日 | 2017-03-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.968 Å) | | 主引用文献 | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
4GK5
 
 | | Crystal structure of human Rev3-Rev7-Rev1-Polkappa complex | | 分子名称: | DNA polymerase kappa, DNA polymerase zeta catalytic subunit, DNA repair protein REV1, ... | | 著者 | Tao, J, Min, X, Wei, X. | | 登録日 | 2012-08-10 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Structural insights into the assembly of human translesion polymerase complexes
Protein Cell, 3, 2012
|
|
4GK0
 
 | | Crystal structure of human Rev3-Rev7-Rev1 complex | | 分子名称: | DNA polymerase zeta catalytic subunit, DNA repair protein REV1, Mitotic spindle assembly checkpoint protein MAD2B | | 著者 | Tao, J, Min, X, Wei, X. | | 登録日 | 2012-08-10 | | 公開日 | 2013-03-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural insights into the assembly of human translesion polymerase complexes
Protein Cell, 3, 2012
|
|
